Polypeptide probe for recognizing G-quadruplex and application of polypeptide probe in detection of G-quadruplex in cells
A quadruplex and probe technology, applied in the field of biological probes, can solve the problems of inability to detect G-quadruplexes, inability to detect G-quadruplexes in cells, etc., and achieve high affinity and selectivity and simple structure. , the effect of small interaction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Preparation of G4P.
[0043] Insert the G4P gene sequence between the Nde I and EcoR I restriction sites of the pet28b vector, and transform it into BL21-DE3 expression bacteria through plasmids. When the bacteria are cultured to OD 0.5, add 0.8mM final concentration of IPTG, and induce 4 Hours, then protein extraction, purification, protein extraction using Capturem TM His-Tagged Purification Miniprep Kit (TAKARA). The purified protein was stored in a solution: 20 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.1 mM EDTA, 50% glycerol at -20°C.
Embodiment 2
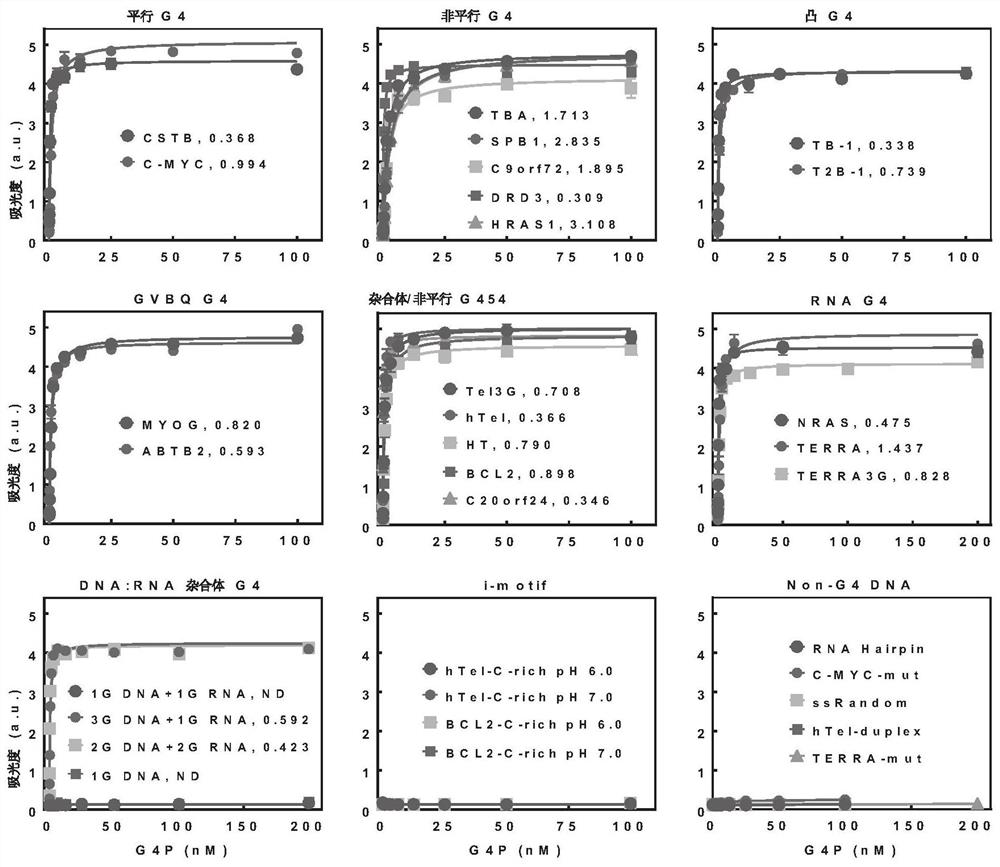
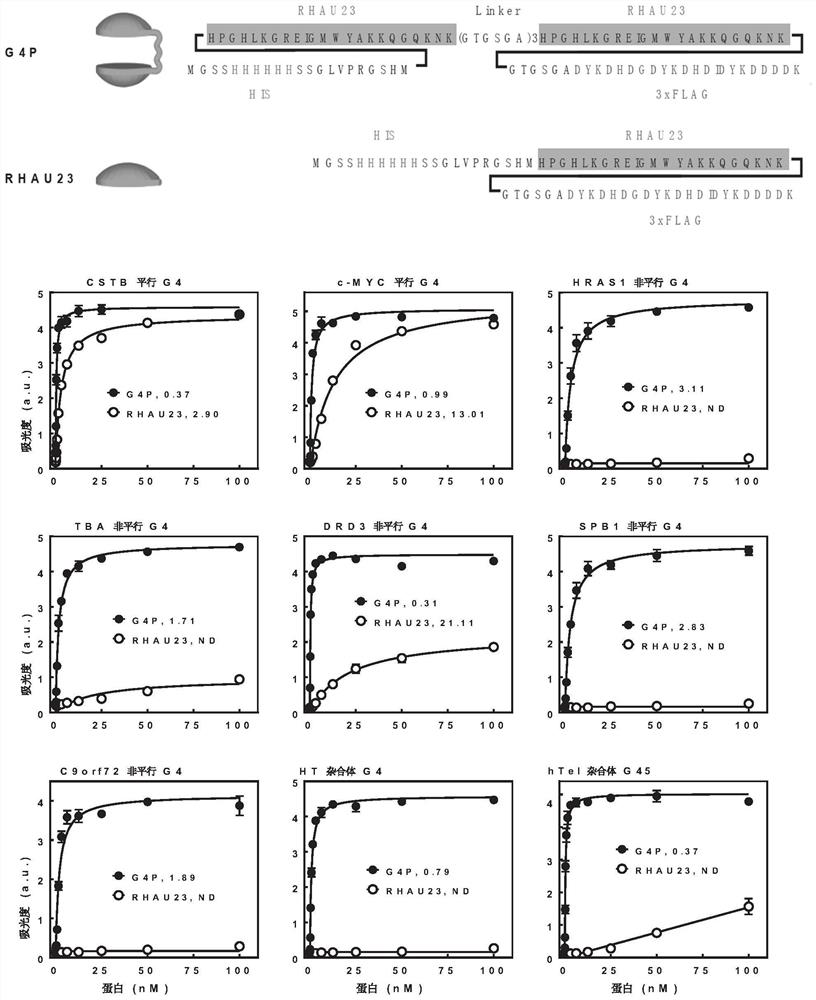
[0045] Validation of the specificity of G4P recognition of G-quadruplexes.
[0046] In this example, the enzyme-linked immunosorbent assay was used to determine the binding ability of G4P to 21 G-quadruplexes of different sequences and several non-G-quadruplex control nucleic acids. The specific method is: heat 21 biotinylated oligonucleotides (see Table 1 for the sequence) in a buffer containing 10mM Tris-HCl (pH 7.4) and 150mM KCl to 95°C, then slowly anneal and cool to 20°C . The annealed oligonucleotides were immobilized on streptavidin-coated multiwell plates (Sigma-Aldrich), and then incubated with corresponding concentrations of G4P or RHAU23. Using anti-FLAG mouse monoclonal antibody (TransgenBiotech, China), HRP-conjugated goat anti-mouse IgG (H+L) secondary antibody (Transgen Biotech, China) and TMBELISA substrate (Transgen Biotech, China), according to the manufacturer's manual to operate. Absorbance at 450 nm was measured on a Multi-PlateReader (Biotek, USA). Dis...
Embodiment 3
[0056] G4P recognizes G-quadruplexes in cells.
[0057] In order to identify G-quadruplexes in cells, the present invention expresses G4P in human A549 cells and uses it to carry out co-immunoprecipitation of G-quadruplexes. After the G4P-bound DNA library is constructed, next-generation sequencing is performed, and then the DNA sequence in the sequencing result is matched to the corresponding site in the genome.
[0058] The main steps are as follows:
[0059] Construction of G4P-ChIP plasmid:
[0060] The DNA encoding G4P was synthesized at Generay Biotechnology (Shanghai, China), and the DNA sequence was inserted between the NheI and EcoRI sites of the pIRES2-EGFP vector to obtain pG4P-IRES2-EGFP. The coding sequence of the nuclear localization signal peptide (NLS: PKKKRKV) of the SV40 major antigen was synthesized in Sangon (Shanghai, China), and inserted into the Nhe I site of pG4P-IRES2-EGFP to obtain the plasmid pNLS-G4P-IRES2-EGFP.
[0061] A DNA fragment expressing...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com