Phosphatidylinositol proteoglycan 3 nanometer antibody as well as preparation method and application thereof
A phosphatidylinositol and nanobody technology, applied in the biological field, can solve the problems of high cost, low antibody stability, and complicated preparation process, and achieve the effects of low cost, high antibody stability, and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1 Preparation of GPC3 recombinant protein
[0037] Prepare GPC3 recombinant protein with FLAG tag at the carboxy-terminus.
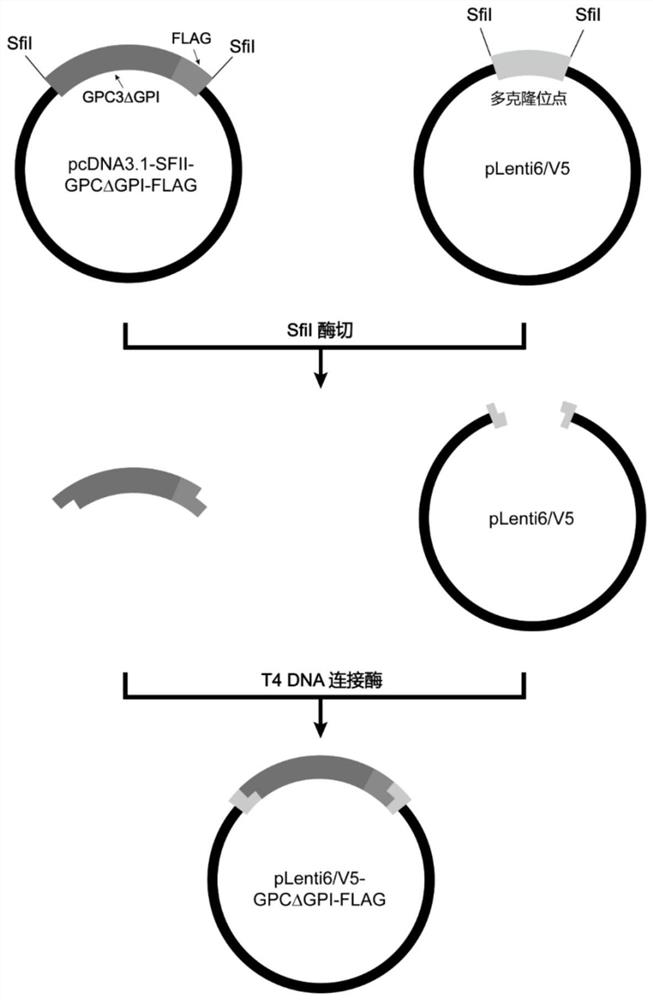
[0038] (1) According to the coding gene sequence of GPC3 in NCBI (NCBI Reference Sequence: NM_004484.4), and based on the amino acid sequence of GPC3 in UniProt (UniProt Reference Sequence: P51654-1), the F359S mutation was performed, and the GPI region ( 1690-1740bp), the fusion sequence of GPC3ΔGPI and FLAG tag missing the GPI site was artificially synthesized (Shenzhen Huada Gene Technology Co., Ltd.), with EcoRI and NotI restriction sites at the 5' end and 3' end, respectively. The amino acid sequence encoded by GPC3ΔGPI is shown in SEQ ID NO:9, and the nucleotide sequence is shown in SEQ ID NO:10. (2) Use restriction endonucleases EcoRI and NotI (New England Biolabs Company) to double the GPC3ΔGPI-FLAG DNA fragment and the pcDNA3.1 vector (intermediate vector) containing SfiI restriction sites at both ends of the modified multiple cl...
Embodiment 2
[0039] Example 2 Screening of Nanobody Phage Library against GPC3
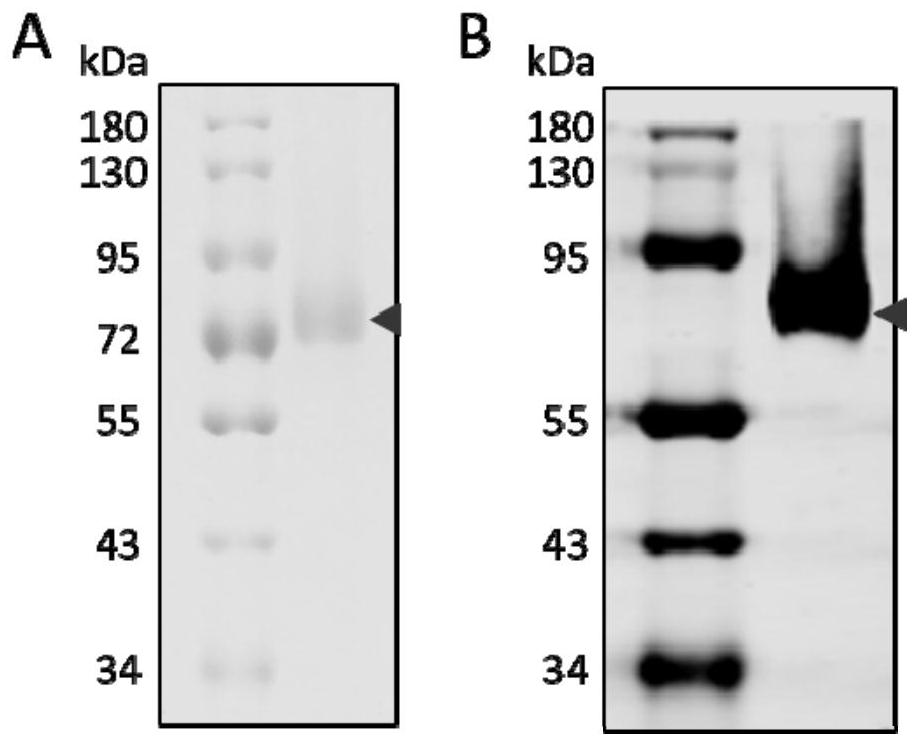
[0040] (1) Coating antigen: According to the instructions of FLAG M2 magnetic beads, the GPC3ΔGPI-FLAG protein secreted into the culture supernatant was purified to prepare GPC3ΔGPI-FLAG protein-coated magnetic beads, which were stained with Coomassie brilliant blue and immunoblotted. Test for verification. Such as figure 2 As shown in (A), the size of the purified protein is between 72-85kD, and the band is single, indicating high purity. figure 2 (B) Western blot detection results using the FLAG tag antibody of the GPC3ΔGPI-FLAG recombinant protein. The results show that the protein size is consistent with the Coomassie brilliant blue staining results, indicating that the purified protein is the target protein of GPC3ΔGPI-FLAG. GPC3 protein has several glycosylation sites, and the trailing diffuse bands should be caused by GPC3 glycosylation. The arrow indicates the target band position of the GPC3ΔGPI-...
Embodiment 3
[0041] Preparation of Example 3 Nanobodies
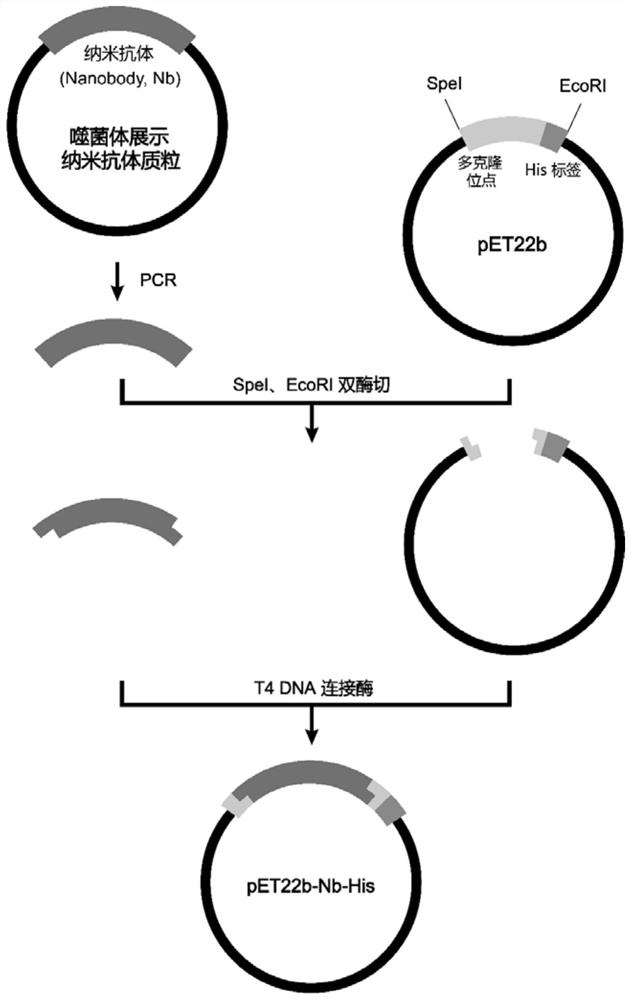
[0042] In Example 2, after completing the third round of screening for phage infection, Escherichia coli SS320 was coated on a plate, and single clones containing phage plasmids were picked for sequencing. The gene sequences of each antibody clone were analyzed and compared using Vector NTI software, and the clones with the same sequence of complementary regions CDR1, CDR2, and CDR3 were regarded as the same clone. According to the sequencing results, one of the clones with a high repetition rate was selected and labeled as G64 clone. The DNA sequence shown is shown in SEQ ID NO:8, and the encoded amino acid sequence is shown in SEQ ID NO:7. The amino acid sequences of CDR1, CDR2, and CDR3 of the three complementary regions are shown in SEQ ID No:1, SEQ ID No:2, and SEQ ID No:3 respectively, and the nucleotide sequences of CDR1, CDR2, and CDR3 are shown in SEQ ID As shown in NO:4, SEQ ID NO:5, and SEQ ID NO:6, the amino acid sequen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com