Double-label joint design method and preparation method thereof
A technology of tags and adapters, which is applied in the field of design and preparation of double-labeled adapters, can solve problems such as incompatibility with single-end sequencing, increased sequencing time, and increased sequencing costs, so as to avoid false positives, reduce the number of additions, and reduce sequencing costs. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] Example 1. Design of dual-sample index adapter and its application to sequencing library construction
[0056] 1. Two-sample index adapter
[0057] 1. Preparation of two-sample index adapter and its library construction kit
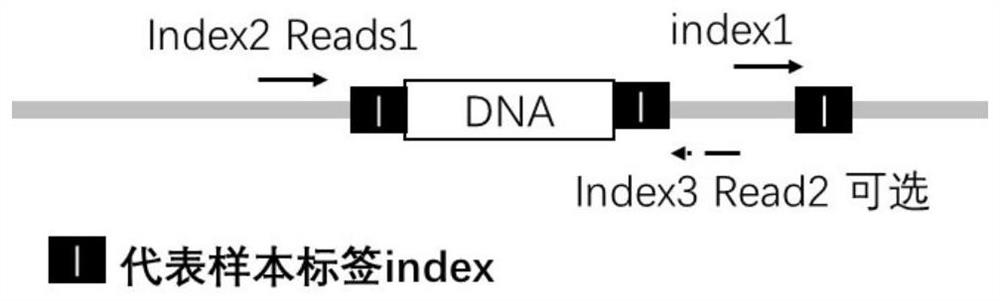
[0058] The second sample index in the two-sample index adapter is located between the sequencing primer binding region and the inserted DNA fragment.
[0059] image 3 The structure of the two-sample index adapter shown in A is as follows:
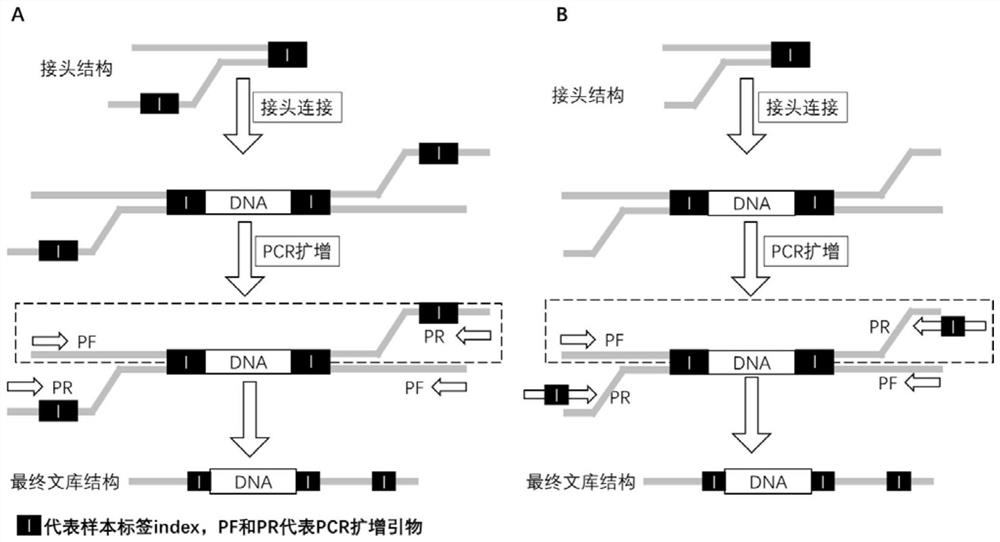
[0060] The double-sample index adapter A is a Y-shaped adapter formed by the annealing of the adapter sequence L-A and the adapter sequence S-A,
[0061] One end of the double-sample index adapter A forms a complementary blunt end or a complementary cohesive end from the complementary region of the adapter sequence L-A and the adapter sequence S-A, and the other end forms a free non-complementary double strand from the non-complementary region of the adapter sequence L-A and the adapter sequence S-A; and the...
Embodiment 2
[0107] Example 2. Design of dual-sample index adapter and its application to sequencing library construction
[0108] 1. Construction of two-sample index adapters
[0109] 1. Construction of double-sample tag adapter sequences
[0110] According to the design scheme of Example 1, design the linker sequence required for constructing the double-sample tag adapter B as shown in Table 1 below;
[0111] The linker sequence in the table was synthesized from Beijing Liuhe Huada Gene Technology Co., Ltd., the purification method was C18 DSL, and the order quantity was 5OD.
[0112] Table 1 Sequences and amplification primer information required to construct adapters
[0113]
[0114] After adapter sequences are ordered, dissolve the master solution to 100 μM in TE buffer.
[0115] 2. Preparation of two-sample index adapters
[0116] Ad01L and Ad01S were mixed according to the same amount of substances, prepared with TE buffer to make a 25 μM joint mixture, left at room temperat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com