Method and device for identifying tumor purity and absolute copy number based on sequencing data
A technology for sequencing data and copy number, applied in the field of tumor research, can solve the problems of increasing the complexity of tumor sample purity information, manual verification, etc., to save labor costs, ensure accuracy, and avoid cumbersome processes.
Active Publication Date: 2020-10-09
深圳吉因加医学检验实验室
View PDF12 Cites 11 Cited by
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
For example, purity estimation software such as ABSOLUTE can predict tumor purity from low-coverage whole-genome sequencing data samples, and is currently one of the most commonly used methods for evaluating tumor purity; its method uses the CNV information of tumor samples to estimate tumor purity. However, due to the complexity of tumor samples, the method also needs to combine SNV information to estimate the tumor purity to achieve higher accuracy. The software will automatically base the predesigned cancer karyotype, somatic mutation frequency and Somatic cell copy number changes are scored and the model with the highest score is selected; however, the model with the highest score is not necessarily the optimal model, which requires manual verification of the results, increasing the complexity of obtaining tumor sample purity information
Method used
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
View moreImage
Smart Image Click on the blue labels to locate them in the text.
Smart ImageViewing Examples
Examples
Experimental program
Comparison scheme
Effect test
Embodiment
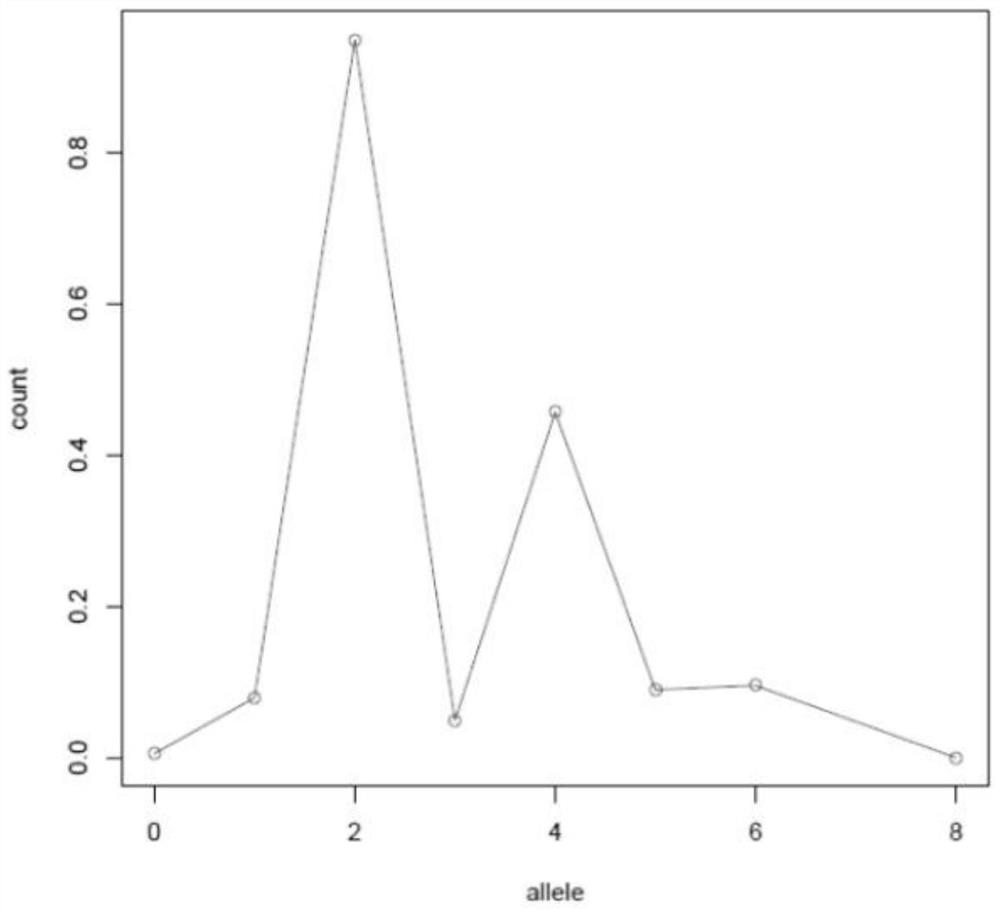
[0093] In this case, for a case of melanoma, numbered 179008702TD, the exome data of normal and tumor tissues were obtained, and the purity and absolute copy number of the melanoma tumor were identified based on the sequencing data, as follows:
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More PUM
 Login to View More
Login to View More Abstract
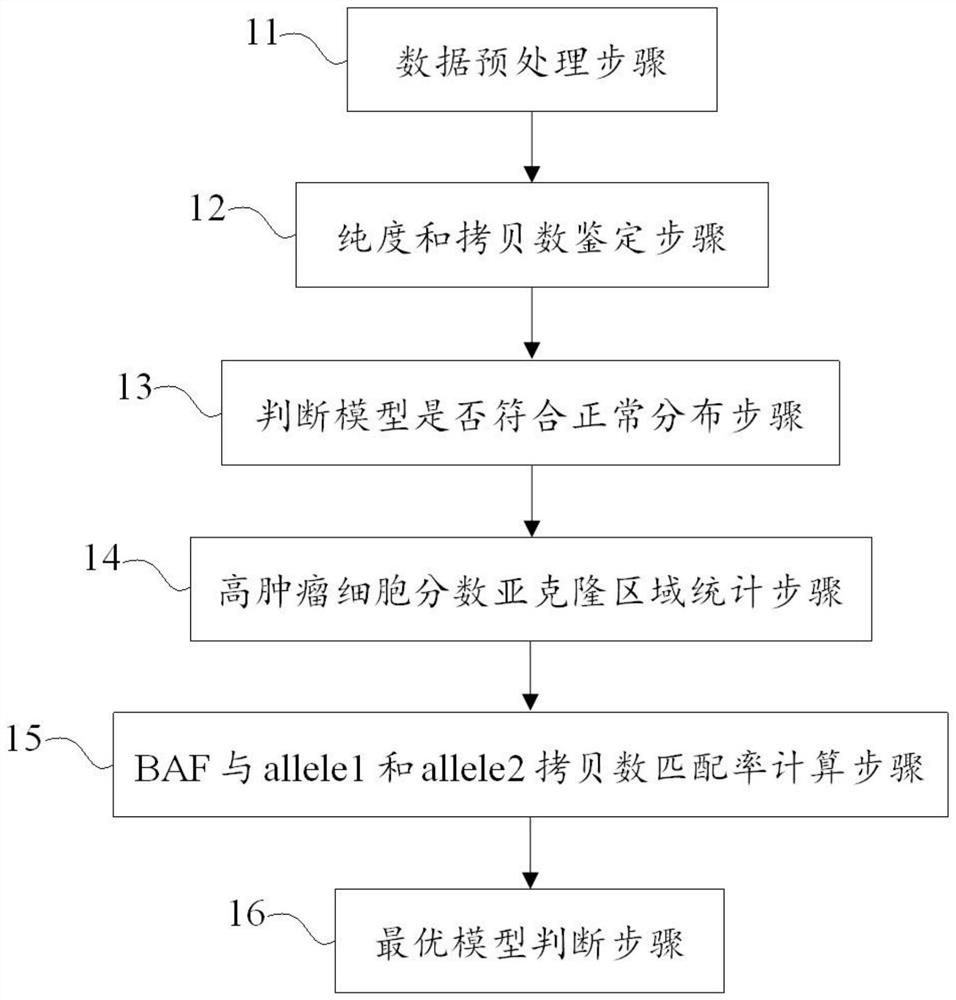
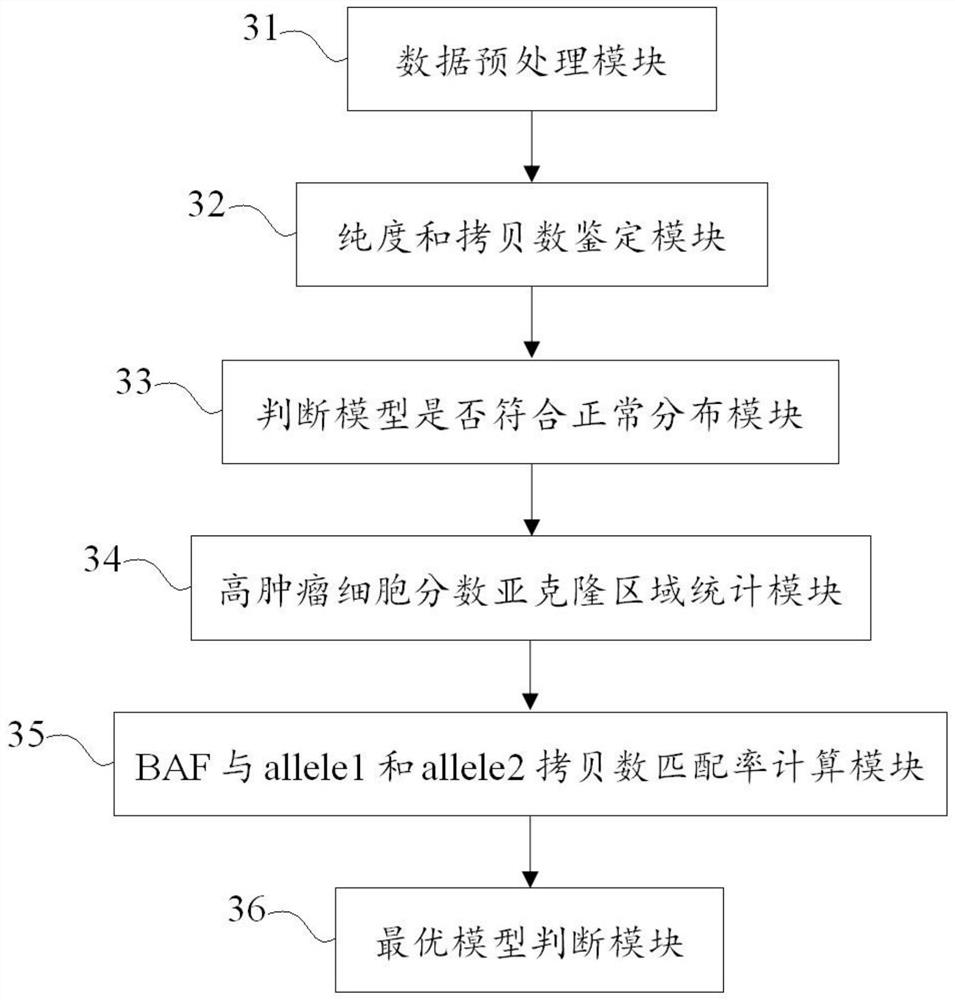
The invention discloses a method and a device for identifying tumor purity and absolute copy number based on sequencing data. The method comprises the following steps: comparing offline data after quality control to a reference genome, and carrying out variation detection and population database annotation; testing the preprocessed data of the tumor and the normal sample by using purity predictionsoftware to obtain a purity and copy number information model; and for the model conforming to normal distribution, further screening out the model with the maximum probe support number in the high tumor cell fraction subcloning region, and defining an optimal model in combination with the matching rate of BAF and alle1 and alle2 copy numbers. According to the method, the model of purity detection software is rapidly and efficiently corrected, and the purity and absolute copy number information of tumors can be obtained more accurately; the accuracy is guaranteed, the tedious process of manual verification is avoided, the labor cost is saved, and a foundation is laid for follow-up tumor genome evolution and tumor heterogeneity research.
Description
technical field [0001] The present application relates to the technical field of tumor research, in particular to a method and device for identifying tumor purity and absolute copy number based on sequencing data. Background technique [0002] Estimating tumor purity and ploidy facilitates studies of tumor genome evolution and intratumoral heterogeneity. The occurrence of tumors is accompanied by a large number of genomic variations, and the definition of chromosome copy number and allele ratio is the basis for understanding the structure and history of tumor genomes. Current genomic identification techniques measure somatic changes in tumor samples in genomic units, ie, DNA quality, and the significance of this measurement depends on the purity and overall ploidy of the tumor. Tumor purity affects the calculation of DNA copy number variation (CNV) to some extent, and ideally, copy number should be measured in copies per tumor cell. Measurement of somatic copy number alter...
Claims
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More Application Information
Patent Timeline
 Login to View More
Login to View More Patent Type & Authority Applications(China)
IPC IPC(8): G16B20/20G16B20/30G16B30/00
CPCG16B20/20G16B20/30G16B30/00
Inventor 黄毅杨玲罗梓文裴士美易鑫刘久成吴玲清李俊刘青峰林浩翔
Owner 深圳吉因加医学检验实验室
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com