Single-gene disease name recommendation method and system based on clinical features and sequence variations
A technology of clinical features and monogenic diseases, applied in the field of medical information, can solve problems such as rare, difficult to understand the phenotype of monogenic diseases, strong heterogeneity of monogenic disease phenotypes, etc., and achieve the effect of accurate diagnosis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
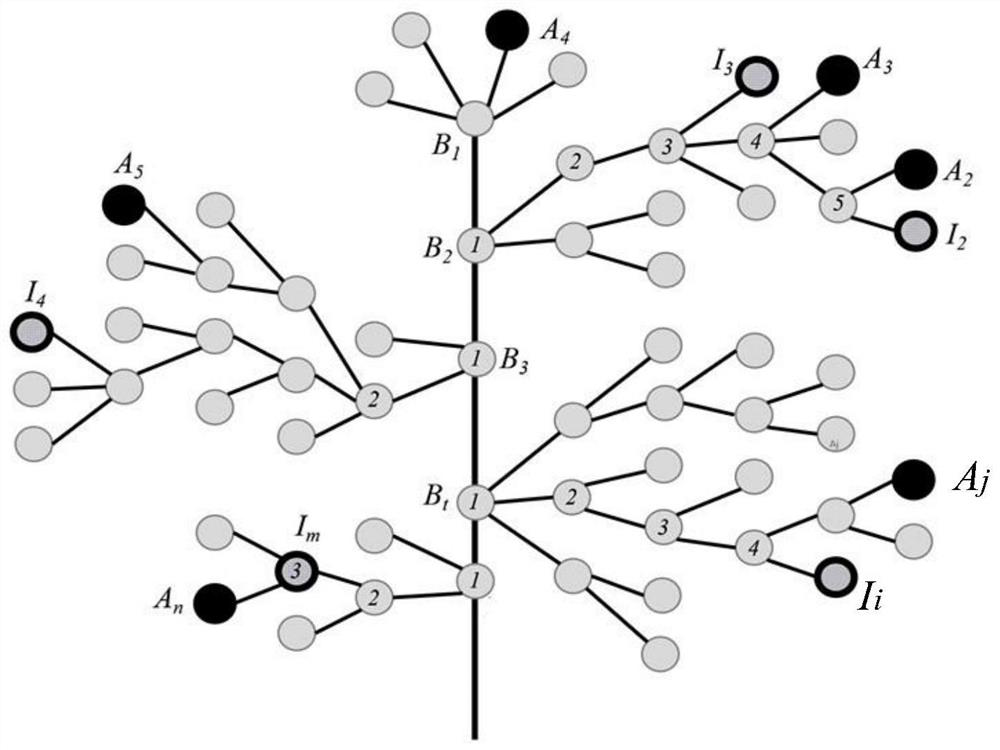
[0068] see figure 1 , this embodiment provides a single-gene disease name recommendation method based on clinical features and sequence variations, including:
[0069] Obtain the patient's case information, which includes gene sequence, feature set I and the name of the monogenic disease; compare the gene sequence with the human reference genome to obtain comparison data, and obtain the impact score of each genetic variation based on the comparison data Traverse the feature set A corresponding to each standard monogenic disease name in the feature relational database, calculate the set similarity value of each feature set A and feature set I respectively, and divide the similar standard monogenic disease names and Corresponding genes are output in descending order, and the standard monogenic disease names of the candidate outputs are aggregated to construct a standard monogenic disease name set P; multiple genes corresponding to monogenic disease names are obtained from the pr...
Embodiment 2
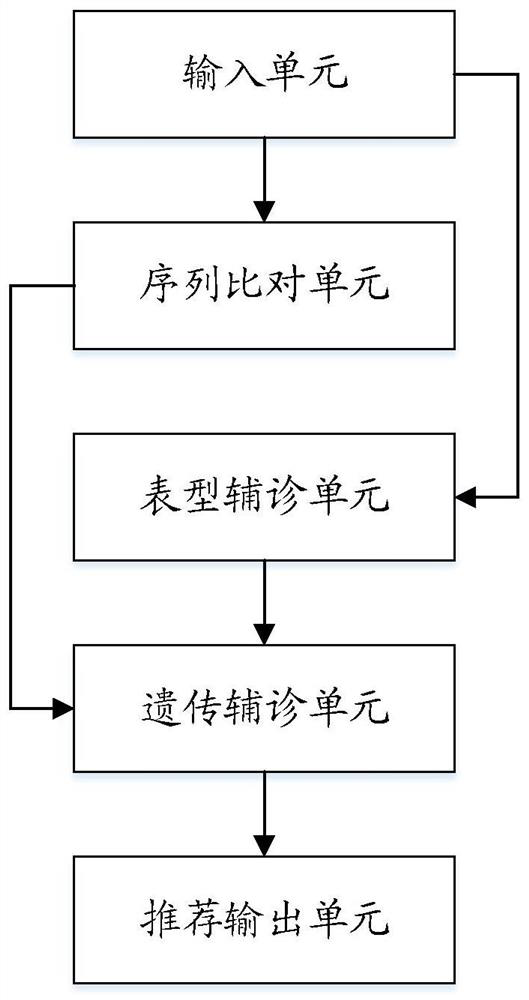
[0189] see image 3 , this embodiment provides a monogenic disease name recommendation system based on clinical features and sequence variations, including:
[0190] The input unit is used to obtain the patient's case information, and the case information includes gene sequence, feature set I and monogenic disease name;
[0191] A sequence comparison unit, which is used to compare the gene sequence with the human reference genome to obtain comparison data, and obtain an impact score for each genetic variation according to the comparison data;
[0192] The phenotype auxiliary diagnosis unit is used to traverse the feature set A corresponding to each standard single-gene disease name in the feature relational database, calculate the set similarity value of each feature set A and feature set I respectively, and compare the similarity values according to the similarity value. The standard single gene disease name and the corresponding gene descending candidate output, and the s...
Embodiment 3
[0197] This embodiment provides a computer-readable storage medium, on which a computer program is stored. When the computer program is run by a processor, the steps of the method for recommending the name of a single-gene disease based on clinical features and sequence variations are executed.
[0198] Compared with the prior art, the beneficial effects of the computer-readable storage medium provided by this embodiment are the same as those of the method for recommending monogenic disease names based on clinical characteristics and sequence variations provided by the above-mentioned technical solution, and details are not repeated here.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com