Application of mir-155 as a marker in the identification or auxiliary identification of animal semen quality
A mir-155, quality technology, applied in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., to achieve the effect of speeding up the breeding process, high accuracy, and improving semen quality traits
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
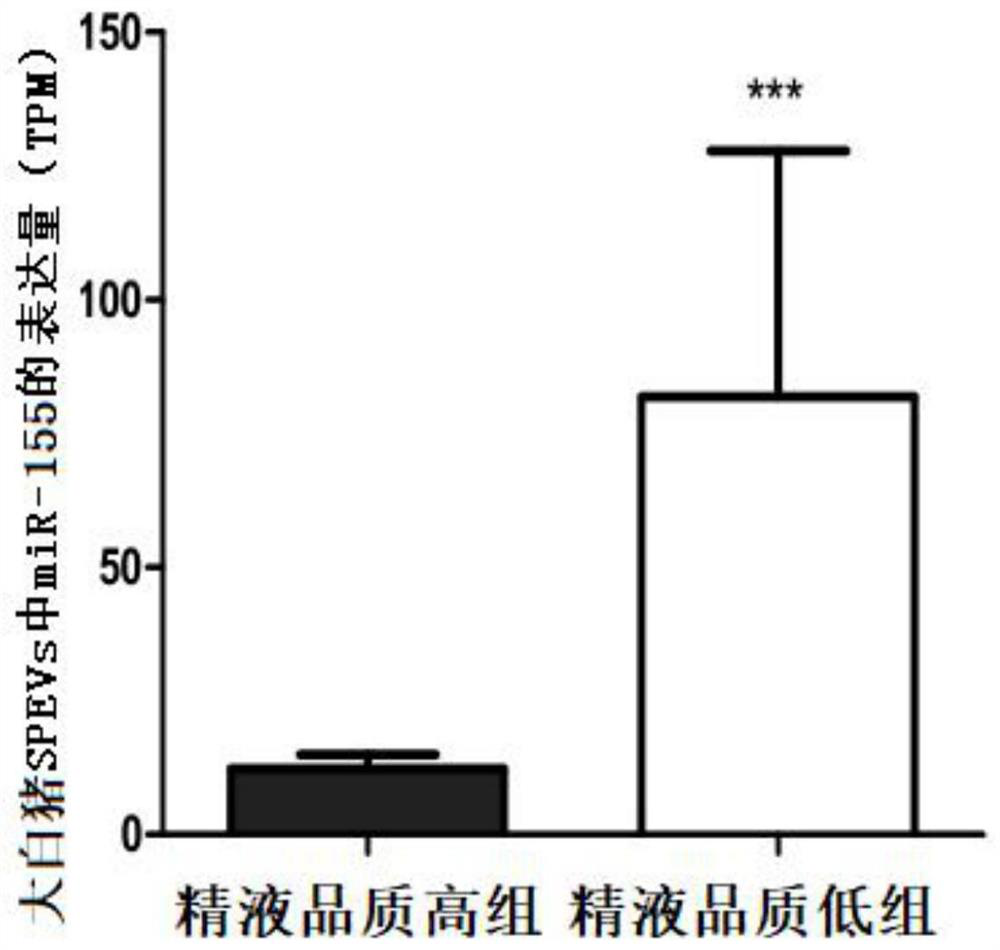
[0068] Example 1. Transcriptome sequencing revealed significant differences in the expression of miR-155 in large white pig seminal extracellular vesicles with high and low semen quality
[0069] 1. Experimental animals and grouping
[0070] The fresh semen samples used in this example are from Henan Jingwang Breeding Pig Improvement Co., Ltd. The boar breed is Large White pig, aged 24-36 months, and raised to sexual maturity under normal nutritional level and feeding conditions. The boars with high semen quality and the boars with low semen quality were screened according to the semen collection records of the boar stud within half a year, and 12 individuals were finally selected. All boars were divided into two groups, a high quality semen group (>90% total motility and >70% fast motility) and a low semen quality group (<80% total motility and <55% fast motility). Fresh semen samples were collected from 12 boars at the same time by semen collectors.
[0071] See Table 1 fo...
Embodiment 2
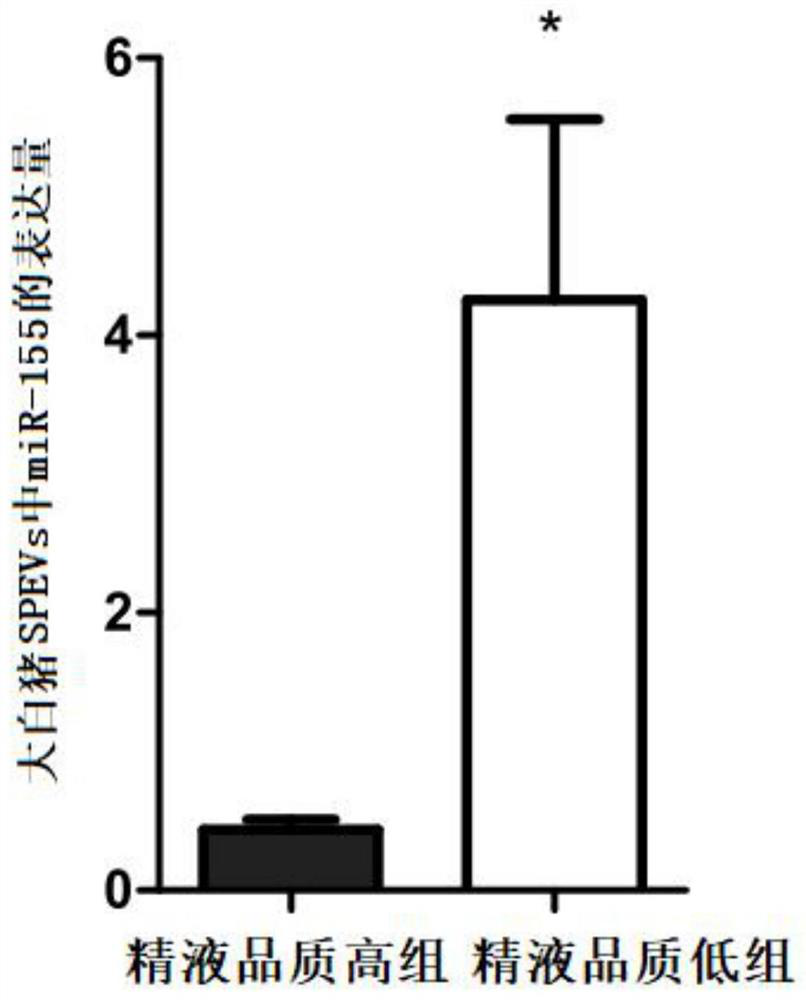
[0113] Example 2. Real-time fluorescent quantitative PCR detection of the expression level of miR-155 in the total RNA (obtained in step 3 in Example 1) of large white pig SPEVs with high and low semen quality
[0114] 1. Preparation of primer pairs
[0115] According to the nucleotide sequence of miR-155, primer pair 1 (composed of primer F1 and primer R1) for reverse transcription and primer pair 2 (composed of primer F2 and primer R2) for real-time fluorescent quantitative PCR detection were designed and synthesized. ).
[0116] The nucleotide sequences of each primer are listed in Table 3.
[0117] table 3
[0118]
[0119] 2. Acquisition of cDNA of porcine SPEVs
[0120] The total RNA of porcine SPEVs obtained in step 3 in Example 1 was respectively taken, and reverse transcription was performed using primer pair 1 to obtain cDNA of porcine SPEVs.
[0121] The reaction system was 15 μL, including 5 μL total RNA of porcine SPEVs, 0.15 μL 100 mM dNTP, 1 μL MultiScrib...
Embodiment 3
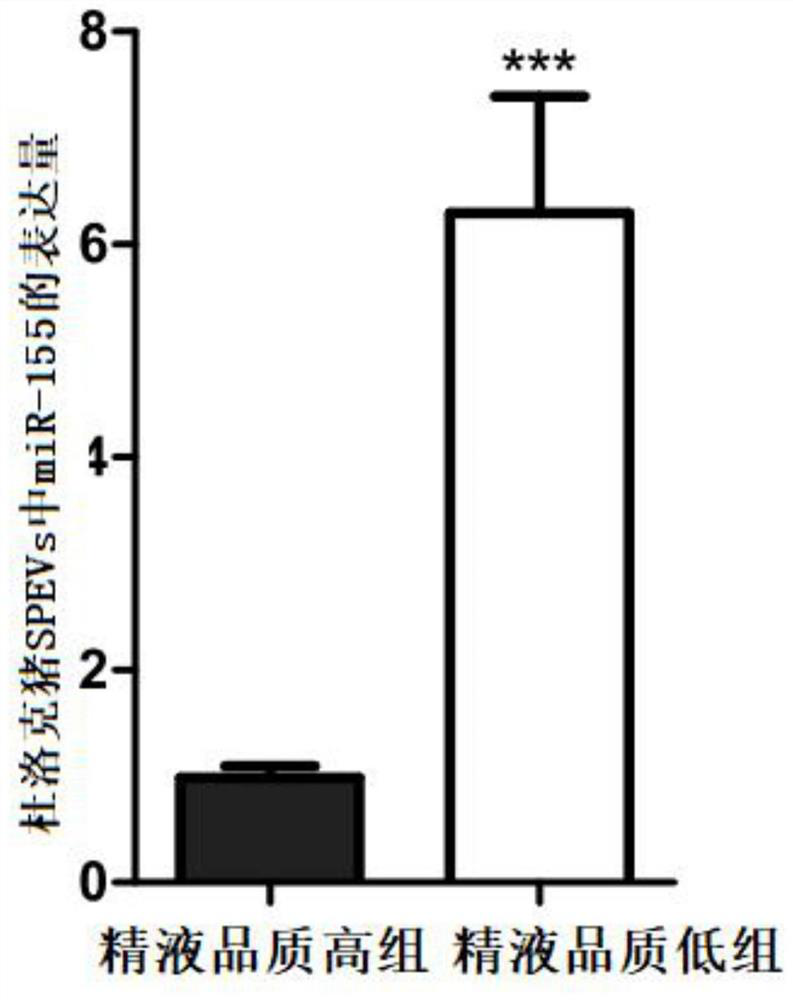
[0135] Example 3. Real-time fluorescent quantitative PCR detection of miR-155 expression in total RNA of Duroc pig SPEVs with high and low semen quality
[0136] 1. Preparation of primer pairs
[0137] Same as Step 1 in Example 2.
[0138] 2. Acquisition of cDNA of porcine SPEVs
[0139] 1. The fresh semen sample used in this example comes from Henan Jingwang Breeding Pig Improvement Co., Ltd. The boar breed is Duroc, the age is 24-36 months, and it is raised to sexual maturity under normal nutritional level and feeding conditions. The boars with high semen quality and the boars with low semen quality were screened according to the semen collection records of the boar stud within half a year, and finally 8 individuals were selected. All boars were divided into two groups, high semen quality group (total motility >90%, fast motility >70% and slow motility 15%). Fresh semen samples were collected from 8 boars at the same time by semen collectors.
[0140] See Table 5 for det...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com