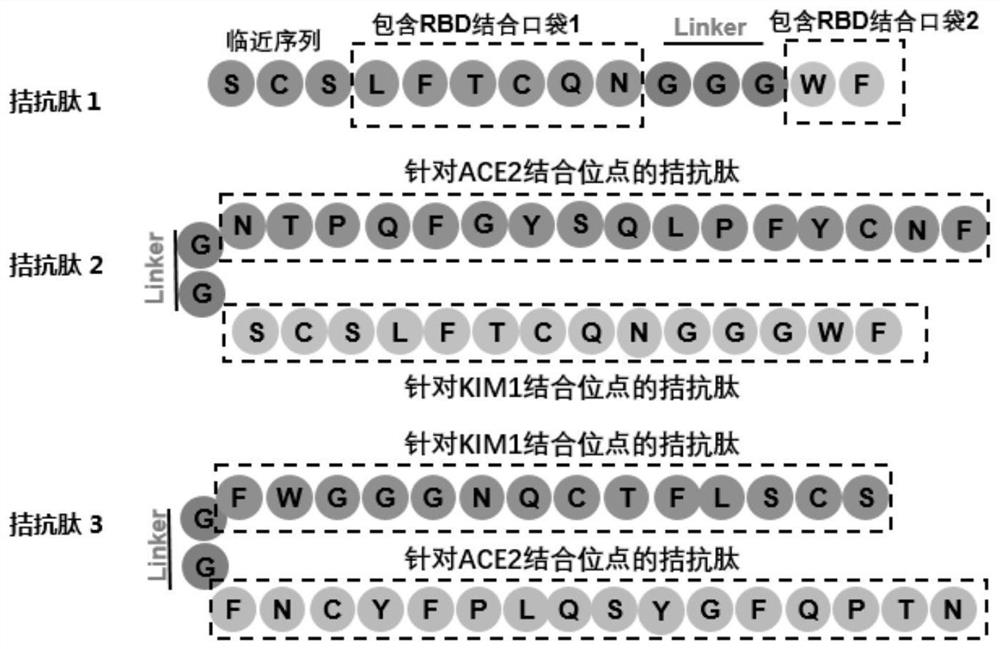
Antagonistic polypeptide and application thereof in preparation of novel coronavirus resistant drugs
A coronavirus, antagonistic technology, applied in the field of biopharmaceuticals, can solve problems such as poor safety, poor efficacy, and lack of drugs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
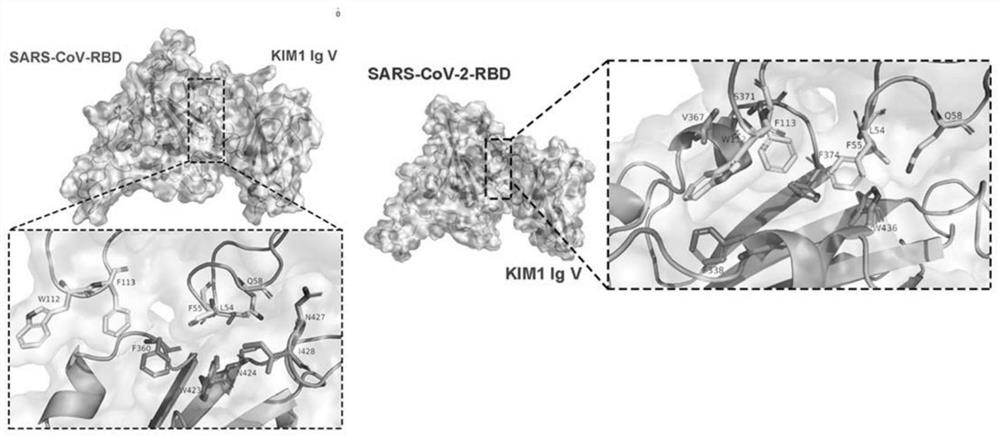
[0038] Leu54, Phe55, Gln58 (binding pocket 1), Trp112, Phe113 (binding pocket 2) of KIM1 can bind to SARS-CoV-2 and SARS-CoV.
[0039] Obtain the crystal structure of KIM1 (PDB ID: 5DZO), SARS-CoV-2-RBD (PDBID: 6M0J) and SARS-CoV-RBD (PBD ID: 2AJF) from the Protein Data Bank database, import the Z-Dock program, find Potential binding pattern diagram. Kinetic analyzes were performed for preferred binding models. Submit 50ns kinetic calculation to study the kinetic parameters of KIM1 and SARS-CoV-2-RBD protein complex, and analyze the obtained MM-GBSA binding energy parameters. The results of molecular simulation docking showed that Leu54, Phe55, Gln58 (binding pocket 1), Trp112, Phe113 (binding pocket 2) of KIM1 could bind to Phe338, Val367, Ser371, Phe374 and Trp436 of SARS-CoV-2, and the cumulative binding The energy is -35.64kcal / mol; and the obtained MM-GBSA binding energy parameters are analyzed. The results of molecular simulation docking show that Leu54, Phe55, Gln58 ...
Embodiment 2
[0048] Antagonist peptide 1 of the present invention has stronger affinity to SARS-CoV-2-spike protein receptor binding domain (RBD).
[0049] Antagonist peptide 1 was modeled by homology modeling techniques and docked with the receptor binding domain (RBD) of the SARS-CoV-2-spike protein, and the binding energy parameters were analyzed. The results show that the binding energy of antagonistic peptide 1 to the receptor binding domain (RBD) of SARS-CoV-2-spike protein is -6.65kcal / mol, indicating that antagonistic peptide 1 has an effect on the receptor-binding domain of SARS-CoV-2-spike protein. (RBD) has a strong affinity.
Embodiment 3
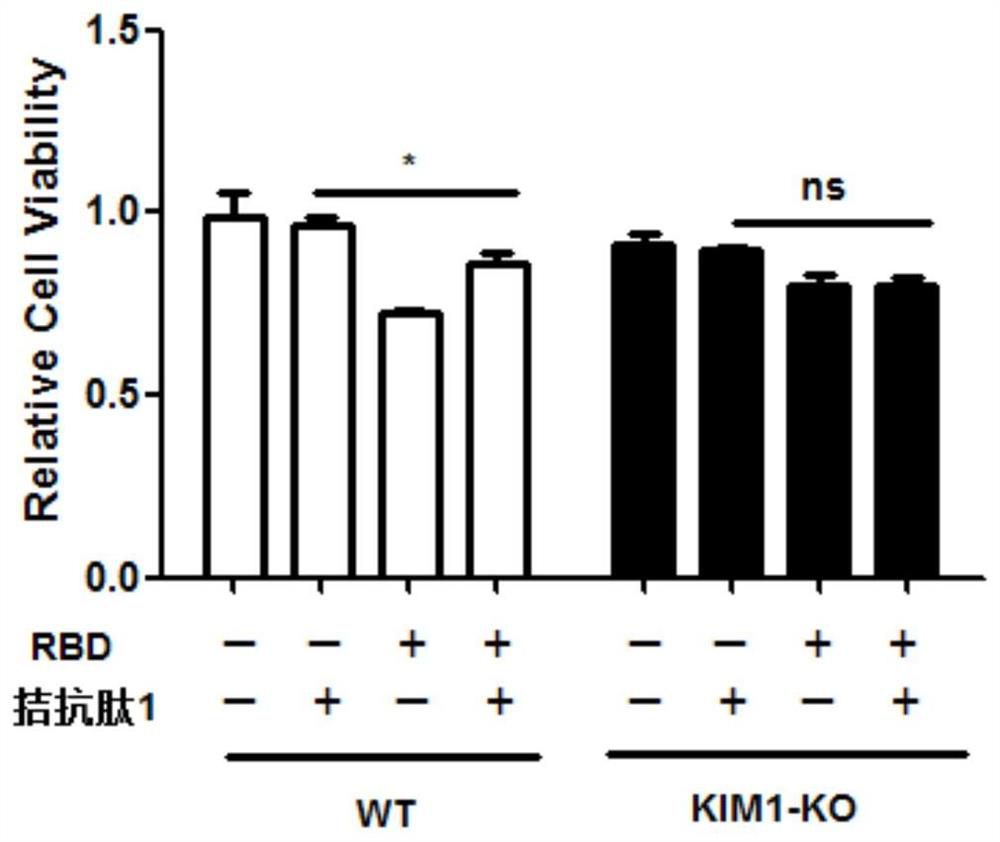
[0051] The antagonistic peptide 1 of the present invention has no obvious cytotoxicity, and can inhibit the cell entry and cytotoxicity of the SARS-CoV-2-spike protein receptor binding domain (RBD).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com