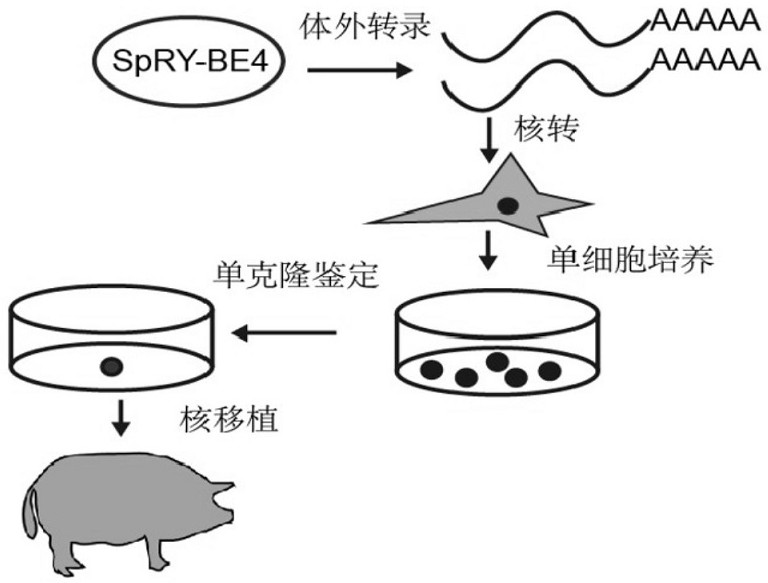
A method for preparing cd163 gene-edited pigs using single base editor spry-be4
A gene editing, rapobec1-xten-cas9n-ugi-nls technology, applied in the biological field, can solve the problems of PAM limitation and no research reports at the level of large animals, so as to avoid random mutation and even genome damage, improve operability and Wideness, effect of increasing freedom and broadness
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] Example 1. Preparation of CD163 biallelic mutant cell lines using a novel single base editor
[0060] 1. Substances for biallelic mutation of CD163
[0061] 1. Expression vector BE4-gRNA
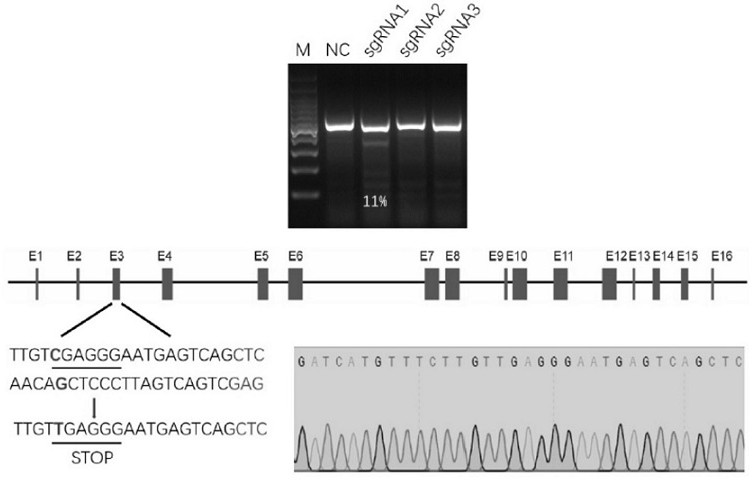
[0062] In this study, the exon 3 region of the CD163 gene was selected as the targeting site. The sequence of the CD163 gene is shown in sequence 1, and the sequence of exon 3 is shown in sequence 2.
[0063] Three gRNAs were designed, and their recognition sequences (target sequences of sgRNAs) were: gRNA-1: TTGTCGAGGGAATGAGTCAG; gRNA-2: TCCCCATCCATCATGTTTGC; gRNA-3: GCAGTCCCAGAGAGCTGACT.
[0064] Synthesize single-stranded oligonucleotides according to the designed sequences, and the synthesis method is as follows:
[0065] gRNA-F1: CACCGTTGTCGAGGGAATGAGTCAG
[0066] gRNA-R1: AAACCTGACTCATTCCCTCGACAAC
[0067] gRNA-F2: CACCGTCCCCATCCATCATGTTTGC
[0068] gRNA-R2: AAACGCAAACATGATGGATGGGGAC
[0069] gRNA-F3: CACCGGCAGTCCCAGAGAGCTGACT
[0070] gRNA-R3: AAACAGTCAGCTCTCTGGGACTGCC ...
Embodiment 2
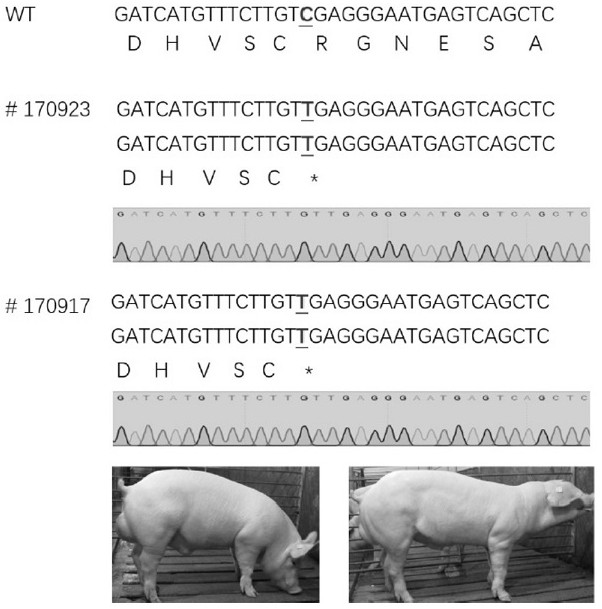
[0160] Embodiment 2, the preparation of CD163 biallelic knockout pig
[0161]1. Preparation of CD163 biallelic knockout pigs
[0162] 1. Take the biallelic C-T mutant fibroblasts in the logarithmic growth phase obtained in Example 1, and digest them with 0.25% trypsin for 5 minutes to obtain single cells, which will become donor cells in subsequent operations.
[0163] 2. Collect the ovaries of adult Large White pigs from the slaughterhouse, wash them three times in PBS solution at 37°C, extract follicles with a diameter of 2-8mm with a needle with a diameter of 0.7mm, and recover cumulus-oocytes with uniform shape and compact structure Cell-complexes (COCs) were washed twice with maturation solution (M199+10%FBS+0.01U / mL bFSH+0.01U / mL bLH+1µg / mL estradiol), and then the cumulus-oocyte- Put 50-60 pieces / well of the complex into a four-well plate containing maturation solution, and place it at 38.5°C, 5% CO 2 After maturing and culturing in the incubator for 18-20 hours, put ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com