Stepped fine adjustment method for in-situ over-expression of long-chain non-coding RNA-LINC00842
An RNA-LINC00842, long-chain non-coding technology, which is applied in the direction of retro-transcriptional RNA virus, DNA/RNA fragments, and other methods of inserting foreign genetic materials, can solve the problem that it is difficult to achieve gradient regulation of target gene expression, and achieve long-term The effect of stabilizing overexpression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
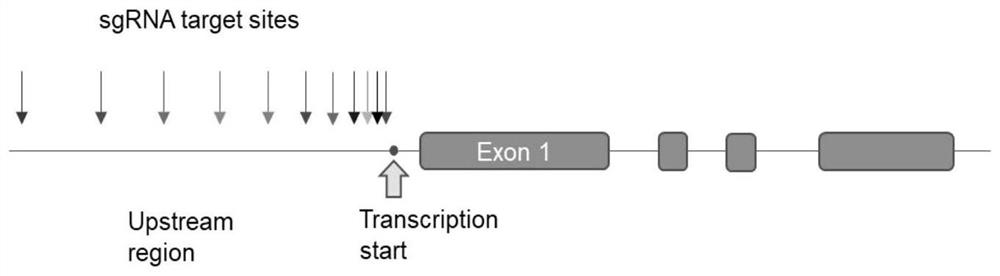
[0041] (1) Basic information of vector system and sgRNA design:
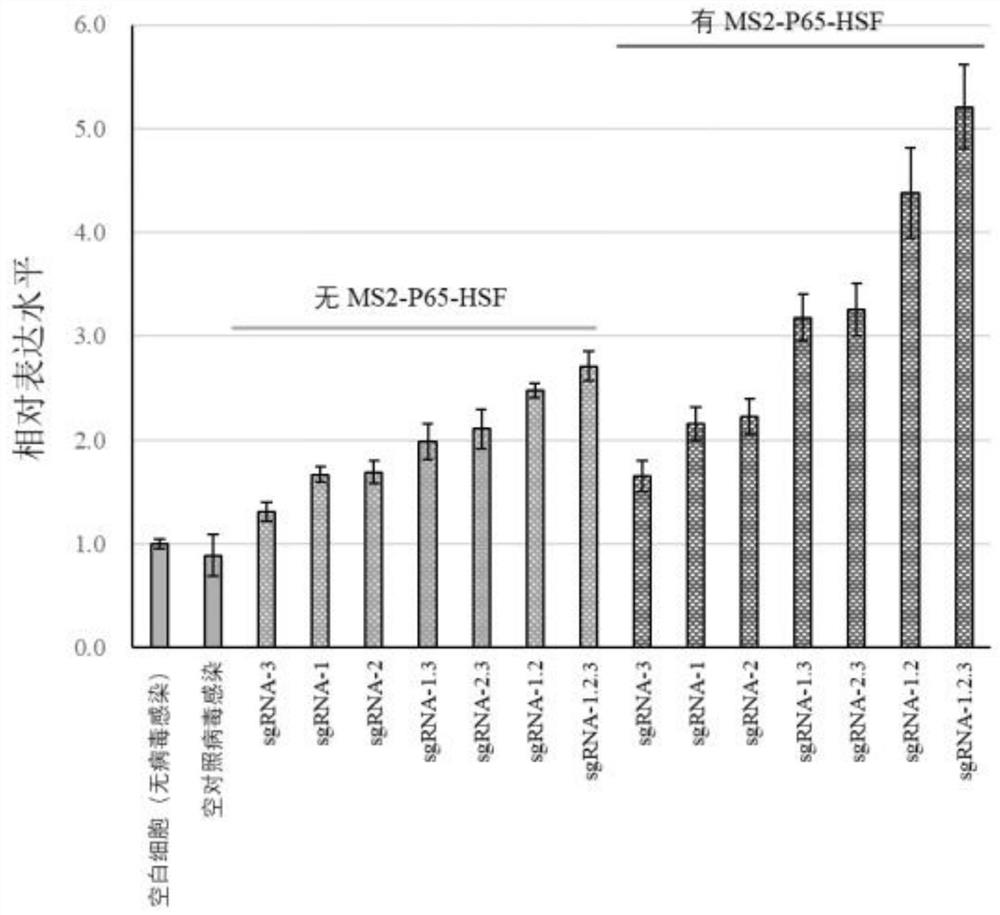
[0042] LINC00842 was overexpressed using the Lenti-CRISPR-dCas9 system, and three sgRNAs were used to achieve gradient regulation of expression levels. Vector construction and lentiviral packaging followed the references (Konermann S, Brigham MD, TrevinoAE, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9complex.Nature 2015;517:583–588), the Lenti-CRISPR-dCas9 system comes from Zhang Feng’s laboratory; the sgRNA sequence is designed using CRISPRdirect.
[0043] Table 1. Lenti-CRISPR-dCas9 system used for overexpression of LINC00842
[0044] Gene Plasmid Mingmu# plasmid name LINC00842 overexpression Plasmid#61425 lentidCAS-VP64_Blast Plasmid#61426 lentiMS2-P65-HSF1_Hygro Plasmid#61427 lentisgRNA(MS2)_zeobackbone
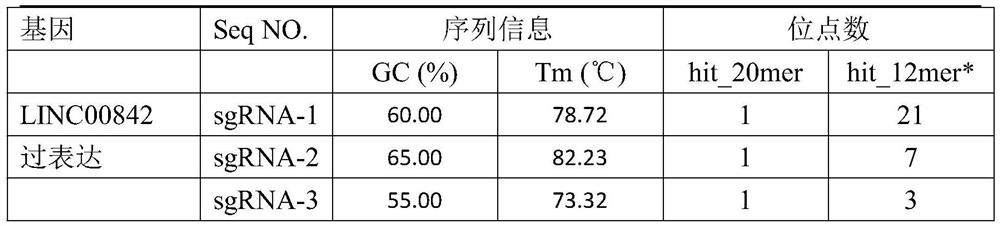
[0045] Table 2. LINC00842 overexpressed sgRNA
[0046] Gene Seq NO. sgRNA sequence LINC00842 overexpression ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com