Screening and application of N-cadherin nucleic acid aptamer based on engineered cells
A nucleic acid aptamer and cell technology, applied in the field of molecular biology, can solve problems such as reduction, loss of EpCAM expression, and reduction of heterogeneous CTCs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
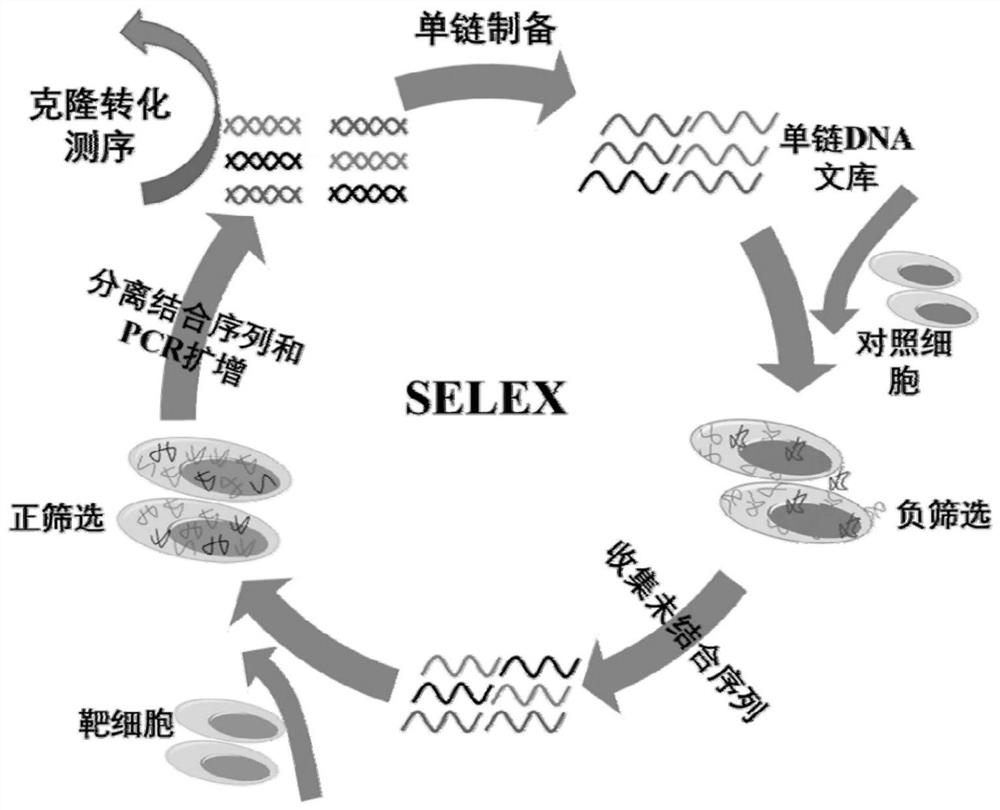
[0067] The screening method for N-cadherin aptamers based on engineered cell lines is as follows:
[0068] a) Using the method of gene amplification, amplify the vector containing the human N-cadherin gene sequence to obtain a large number of N-cadherin sequences, and perform gel electrophoresis to confirm that the amplified sequence is correct and cut it Gel purification; then the purified gene sequence was digested, and the pLVX-IRES-Pruo vector was digested at the same time, and the digested fragments were subjected to electrophoresis, and the target sequence and the digested fragments were confirmed to be correct. The vectors were connected overnight, and then the connected plasmids were cloned and transformed, and then sequenced to verify whether the connected plasmids containing the target sequences were correct and whether the target sequences were correctly connected to the corresponding sites of the target vector.
[0069] b) Name the vector containing the target sequ...
Embodiment 2
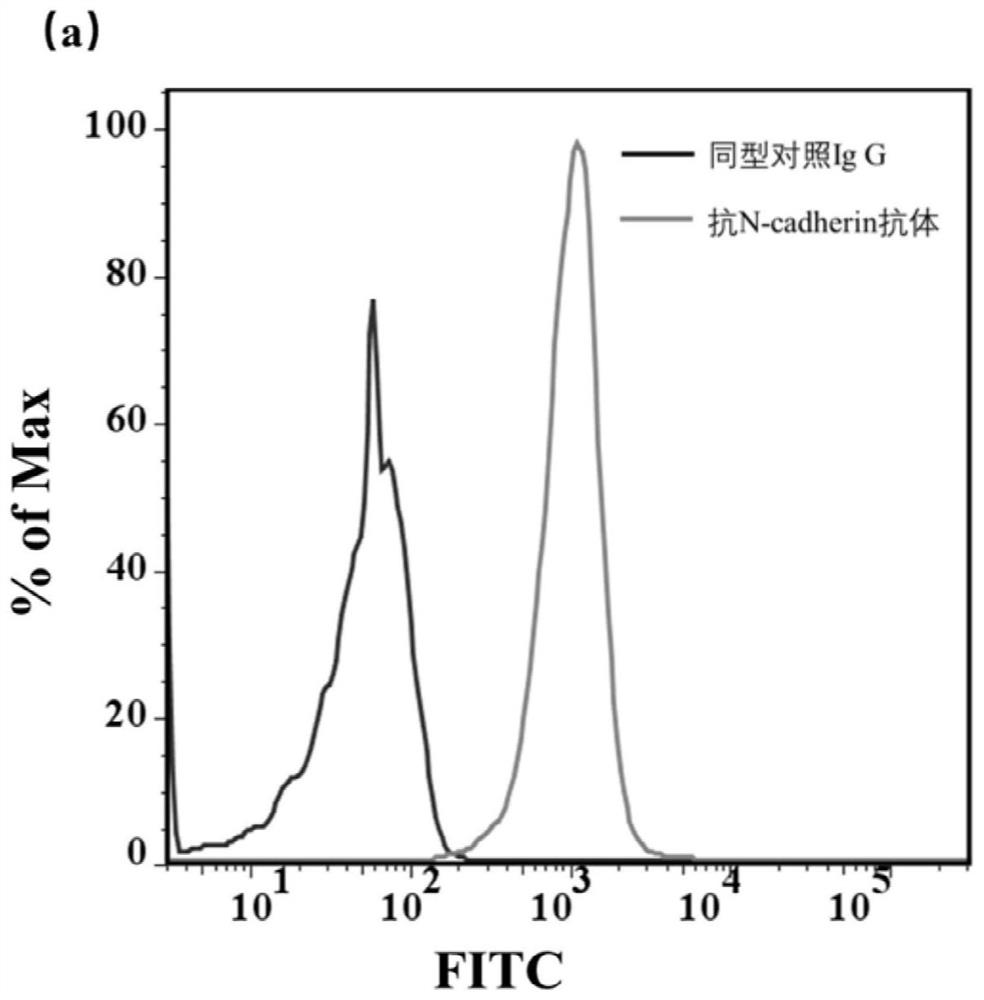
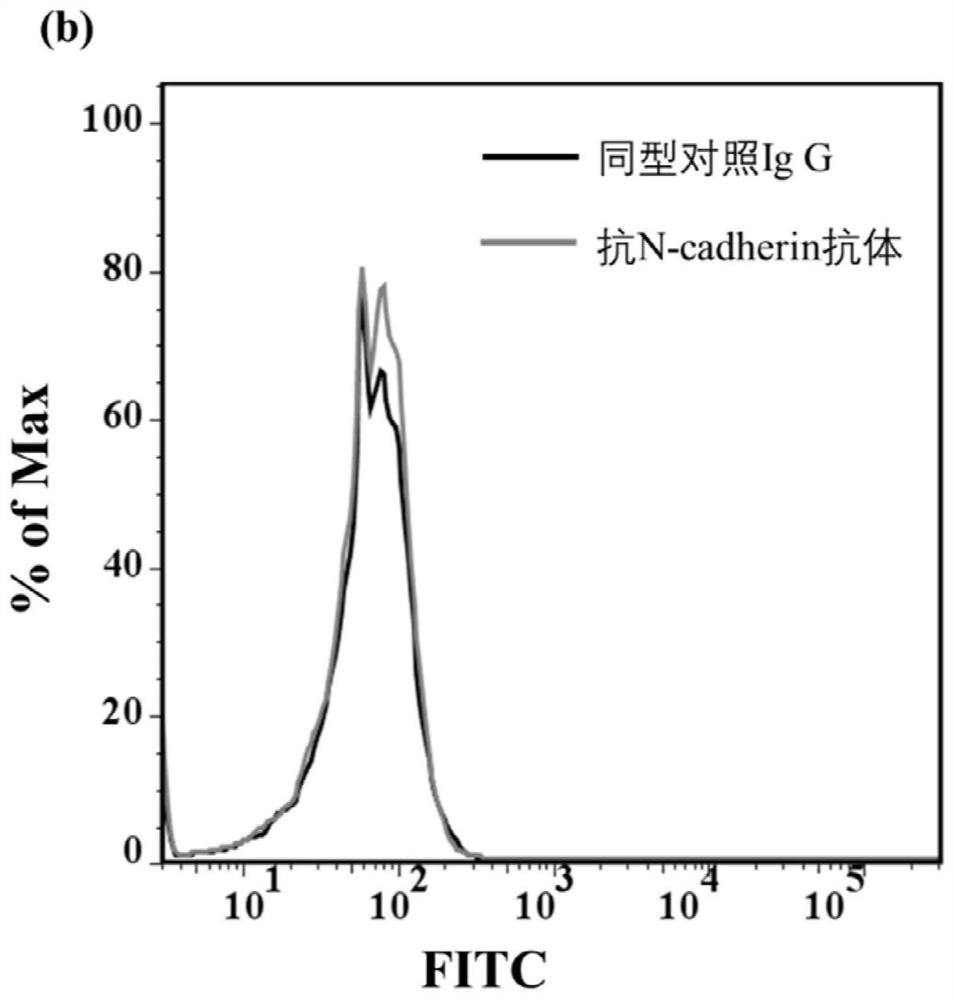
[0073] N-cadherin cells were used as positive cells, and untransfected CHO / K1 cells were used as control cells for aptamer screening. The specific screening method is as follows: first, 10nmol of the designed screening library m-lib was dissolved in 500ul sterilized water, then denatured at 95°C for 5min, and then immediately placed on ice for 10min. The N-cadherin cells with a confluence of 90% were washed three times with washing buffer, and then the cells were incubated with the screening library at 4°C for 60 min, and then washed with washing buffer, and the washing was carried out at 4°C. Wash for 3 minutes each time, and wash three times. Finally, the cells were scraped off with a cell scraper with 1ml of sterilized water, and transferred to a centrifuge tube. After being treated at 95°C for 10 minutes, centrifuged, the supernatant collected was the enriched library obtained in this round of screening. Then, the screened sequences were amplified by PCR, and ssDNA was pr...
Embodiment 3
[0075] N-cadherin cells were used as positive cells, and CHO / K1 cell line was used as control cells to verify the screening effect. The affinity effect of the 12th-round screening library and the screening library m-lib to cells were investigated respectively. After digesting the cells in good condition, take a certain number of cells and plant them in a special confocal culture dish. After 24 hours, the well-growing N-cadherin and CHO / K1 cells were washed three times with PBS. Then 500nM m-lib and fluorescence-modified twelfth-round screening library and positive cells and control cell lines were incubated at 4°C for 50min, then washed three times with 700ul washing buffer, and then imaged by confocal microscopy. Figure 4 and Figure 5 The confocal results showed that the enrichment effect of sequences with strong binding ability to target cells was better when the screening reached 12 rounds.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com