Set of adenine base editors as well as related biological materials and application thereof
A technique for biomaterials, editing, applied in applications, biochemical devices and methods, plant products, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
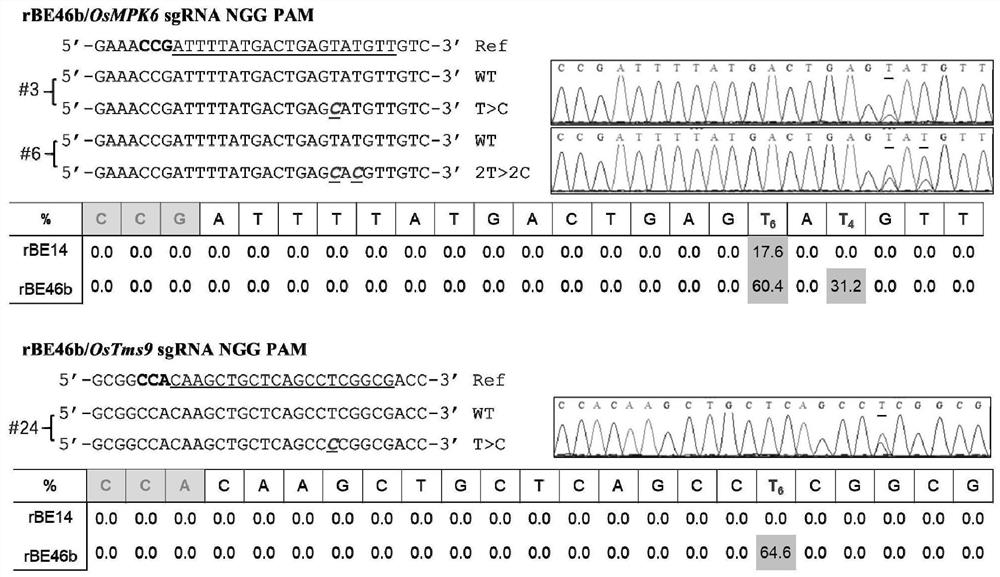
[0043] Example 1. Site-directed mutation of A in the rice genome to G
[0044] 1. Construction of rice adenine base editor expression vector
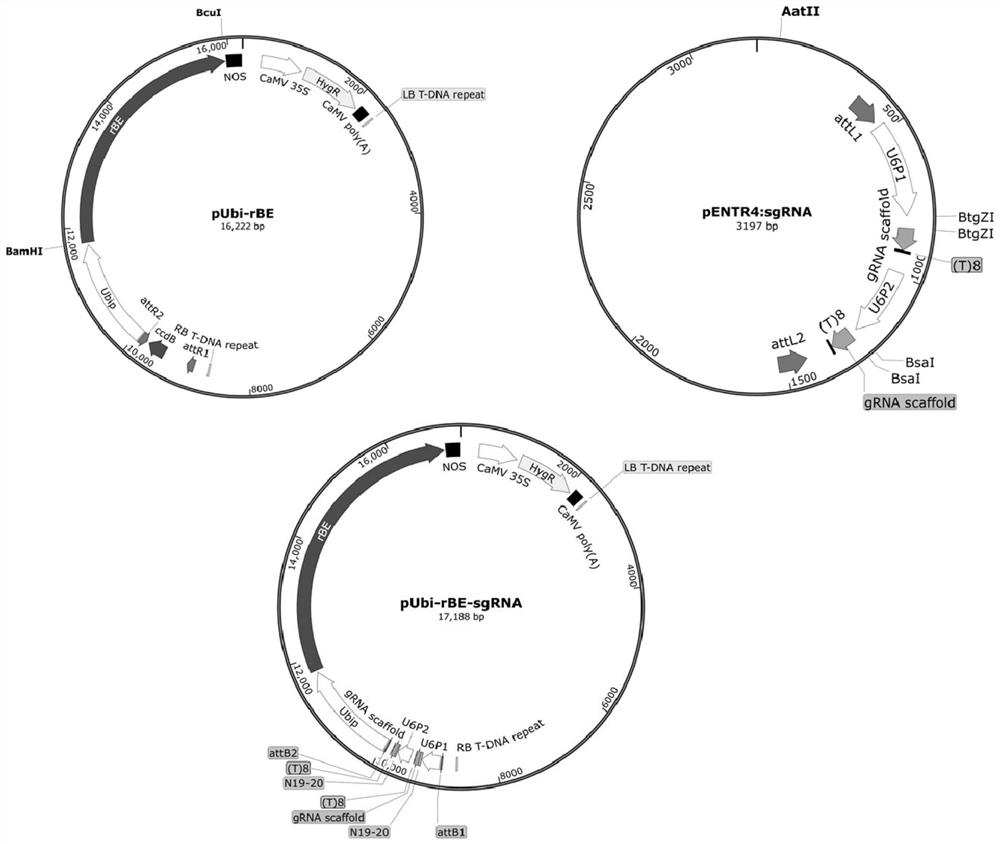
[0045] This embodiment provides four kinds of rice adenine base editor expression vector pUbi-rBE of the present invention ( figure 1 ), named pUbi-rBE46b, pUbi-rBE50, pUbi-rBE54 and pUbi-rBE62, respectively. The adenine base editor expressed by pUbi-rBE46b is a fusion protein named rBE46b (also known as TadA-R-SpCas9(D10A)), composed of adenine deaminase named TadA-R and SpCas9(D10A) The Cas protein is connected with the nuclear localization signal named NLS. The amino acid sequence of rBE46b is SEQ ID No.2 in the sequence listing. In SEQ ID No.2, positions 1-167 are the amino acid sequence of TadA-R, positions 168-199 are the amino acid sequence of the connecting peptide, positions 200-1567 are the amino acid sequence of SpCas9 (D10A), and positions 1568-1576 Bit is the amino acid sequence of NLS. According to the rice codon usa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com