Novel application of compound in prevention and/or treatment of diseases caused by coronavirus infection
A coronavirus, disease technology, applied in medicinal chemistry
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1 protein-protein interaction test
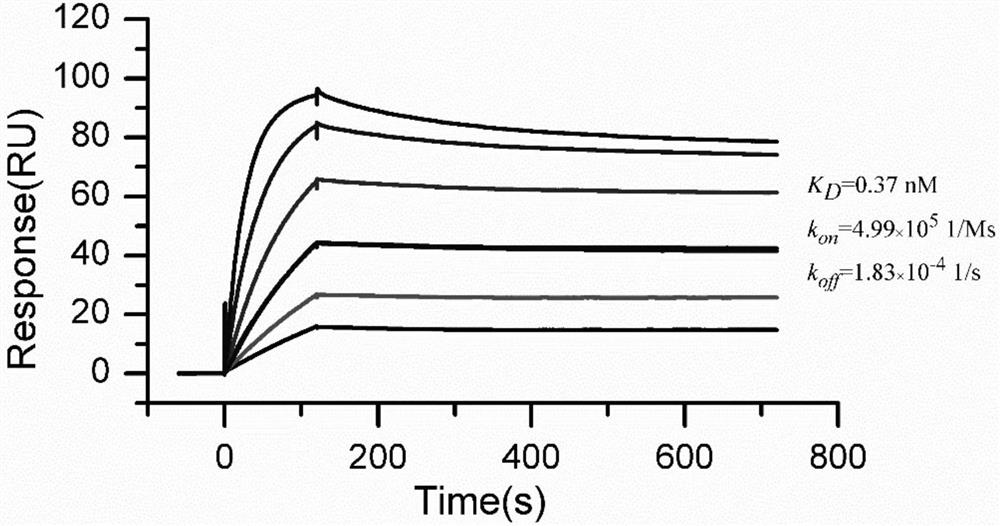
[0035] Protein-protein interactions of ACE2 and RBD proteins were determined using NTA chips using a Biacore S200 instrument. The running buffer was HBS-T+ buffer (10mM HEPES, 150mM NaCl, 0.05% tween 20, pH 8.0, 0.22μm microporous membrane filtration). The ligand is ACE2-His, the concentration is 10 mg / mL, and the capture time is 100 seconds. A series of S-RBD-mFc solutions at concentrations of 3.13, 6.25, 12.5, 25, 50, and 100 nM were prepared, flowed over the captured ACE2-His surface, and the response units (RUs) obtained were recorded. The flow rate was set at 30 μL / min, the binding time was 120 s, and the dissociation time was 500 s. 350mM EDTA is the regeneration solution, and the regeneration time is 60 seconds.
[0036] The experimental results showed that the amount of ACE2 captured by different cycles was relatively stable (the response was about 143RU, and the deviation did not exceed 2RU). In addition, the sen...
Embodiment 2
[0037] Example 2 Establishment of Amino Coupling Method SPR Screening Model
[0038] RBD and ACE2 proteins were immobilized on the CM5 chip by amino coupling method. The chip is first activated by contact with a mixture of EDC and NHS. Then RBD and ACE2-His proteins with concentrations of 20 μg / mL and 16 μg / mL were dissolved in sodium acetate solution (pH 5.0 and pH 4.5, respectively), and flowed over the surface of the chip, and finally immobilized on the Fc2 and ACE2-His of the CM5 chip, respectively. on Fc4. Finally, the chip was blocked with ethanolamine.
[0039] It has been measured that the coupling amount of RBD protein is about 8500RUs, and the coupling amount of ACE2 protein is about 9100RUs. When about 20nM of ACE2 or RBD was passed through the chip immobilized with RBD or ACE2, the response signals were 242RUs and 319RUs, respectively. The above results indicated that both target proteins could maintain their activity after immobilization.
Embodiment 3
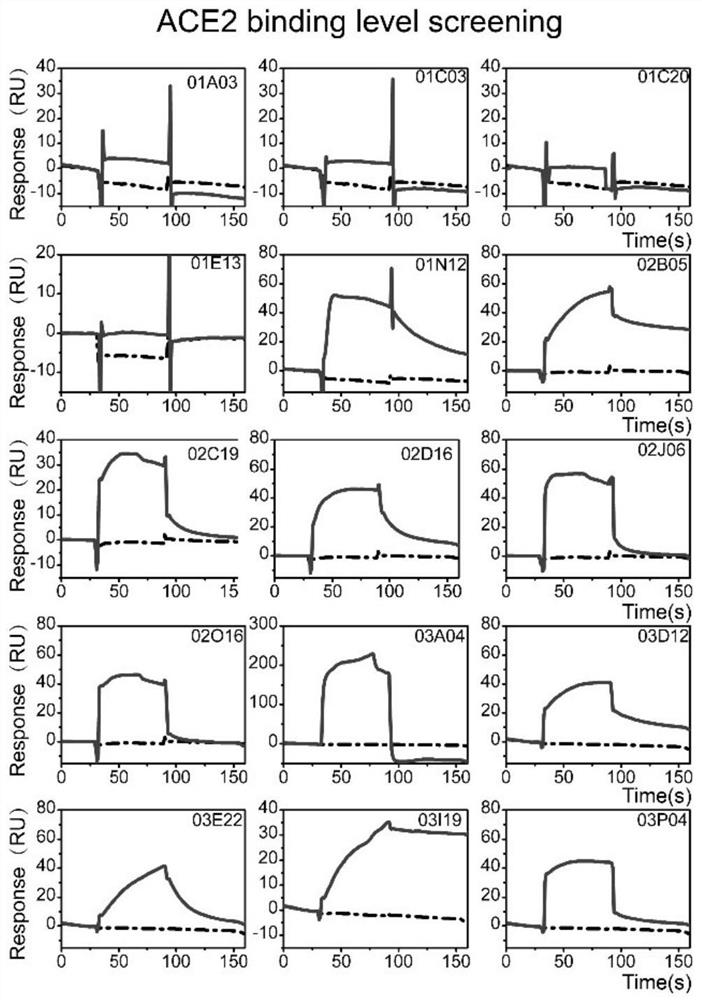
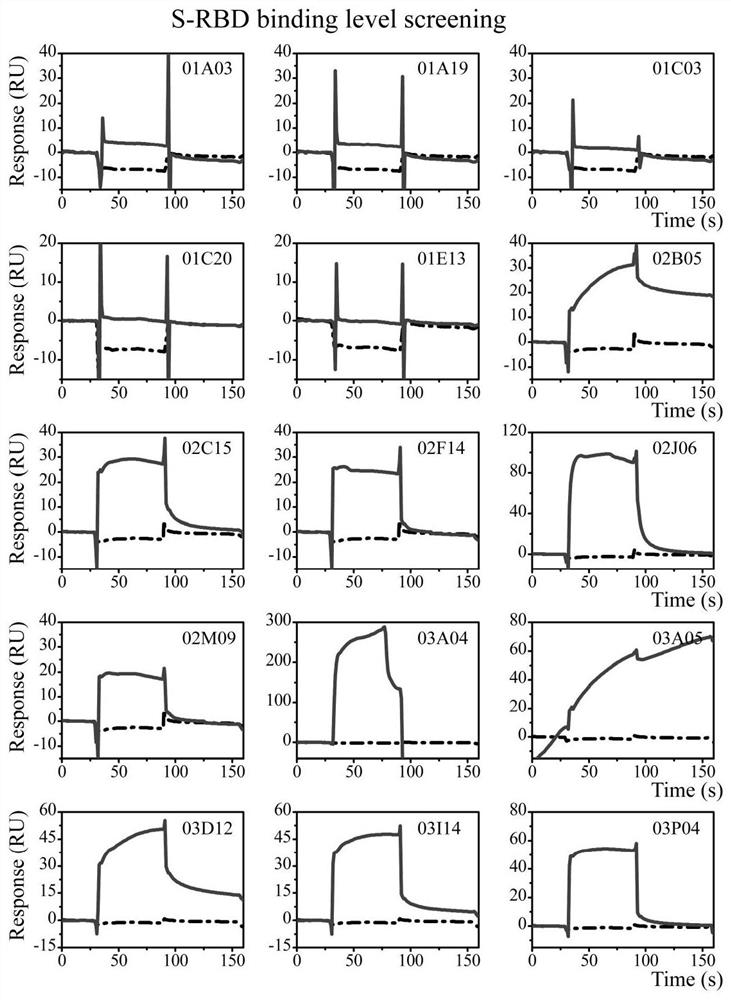
[0040] Embodiment 3 active compound screening
[0041] The whole process of compound screening is divided into three steps: library clearing, binding level screening and affinity determination. All experiments were carried out using Biacore T200 or Biacore S200 instruments with a flow rate of 30 μL / min, and the data were analyzed using Biacore T200 / S200 evaluation software. HBS-T+ buffer (10 mM HEPES, 150 mM NaCl, 0.05% tween 20, 5% DMSO, pH 8.0, 0.22 μm Millipore membrane filtration) was used as the running buffer. Compounds were first diluted to 100 μM and finally diluted to 25 μM or 10 μM in 5% DMSO running buffer.
[0042] Based on the difference in solubility, the library was cleared by flowing the compound at a concentration of 25 μM or 10 μM over the surface of the chip. A total of 13 high-viscosity compounds were excluded by clearing the library. Compounds that pass through the library are then screened for binding levels in order to identify compounds that bind to ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Affinity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com