A protein structure prediction method and device
A protein structure and prediction method technology, which is applied in the field of protein structure prediction based on multi-task time-domain convolutional neural network, can solve the problems of poor robustness and low accuracy, so as to improve the degree of fit, reduce the complexity, improve the The effect of generalization
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0028] The principles and features of the present invention are described below in conjunction with the accompanying drawings, and the examples given are only used to explain the present invention, and are not intended to limit the scope of the present invention.
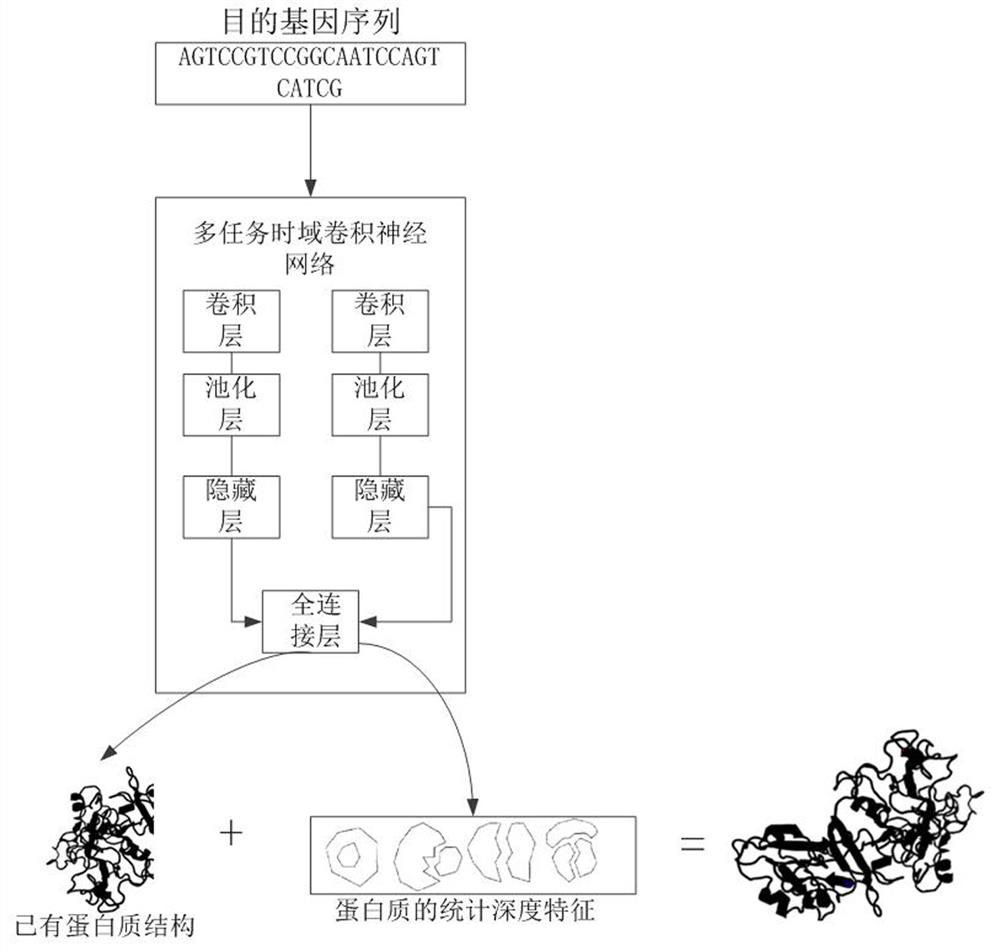
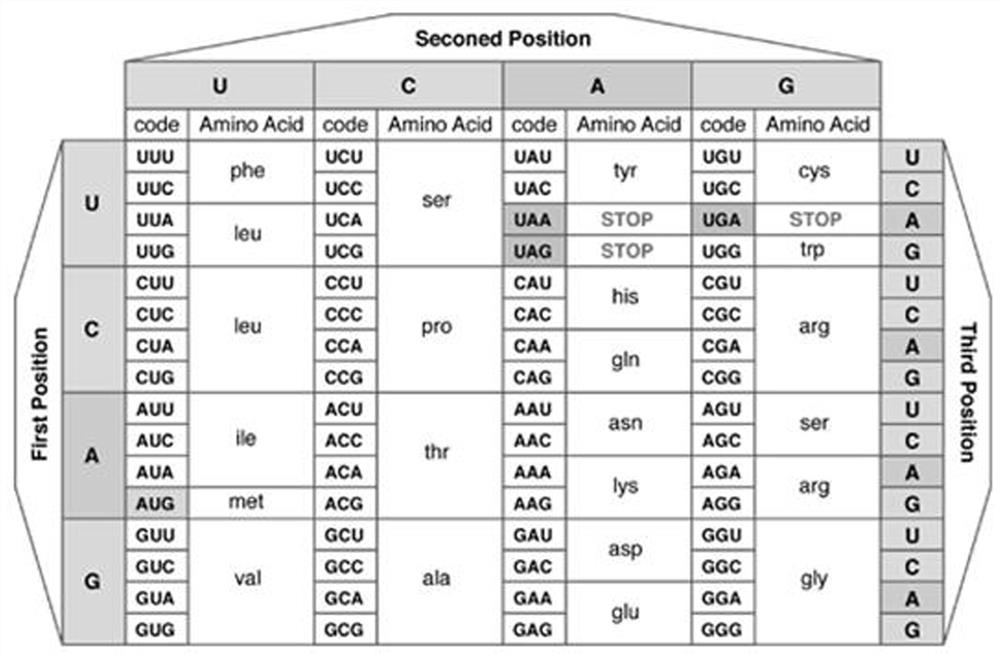
[0029] refer to Figure 1 to Figure 3, in the first aspect of the present invention, a protein structure prediction method based on multi-task time-domain convolutional neural network is provided, comprising the following steps: S101. Obtaining the target gene sequence and protein database; S102. According to the genetic code table and protein The database establishes a DNA-RNA-amino acid triple sequence data set corresponding to each protein; according to the residue depth and physical and chemical properties of the amino acids that make up each protein in the protein database, a multiple regression equation is established to obtain the statistical depth characteristics of each protein ; S103. Clustering the triple...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com