Immunohistochemical nuclear staining section cell positioning multi-domain co-adaptation training method
An immunohistochemistry and cell localization technology, applied in the field of deep learning, which can solve problems such as performance limitations of key point detection models
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0032] Hereinafter, exemplary embodiments of the present application will be described in detail with reference to the accompanying drawings. Apparently, the described embodiments are only some of the embodiments of the present application, rather than all the embodiments of the present application. It should be understood that the present application is not limited by the exemplary embodiments described here.
[0033] Application overview
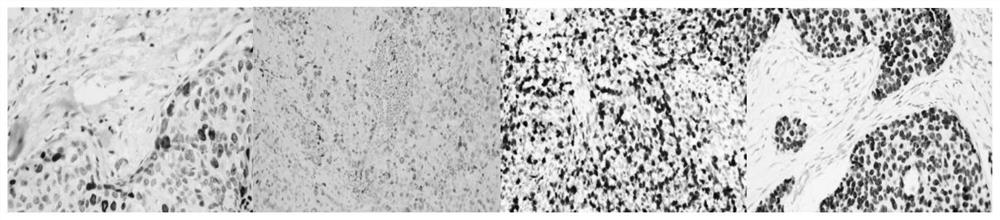
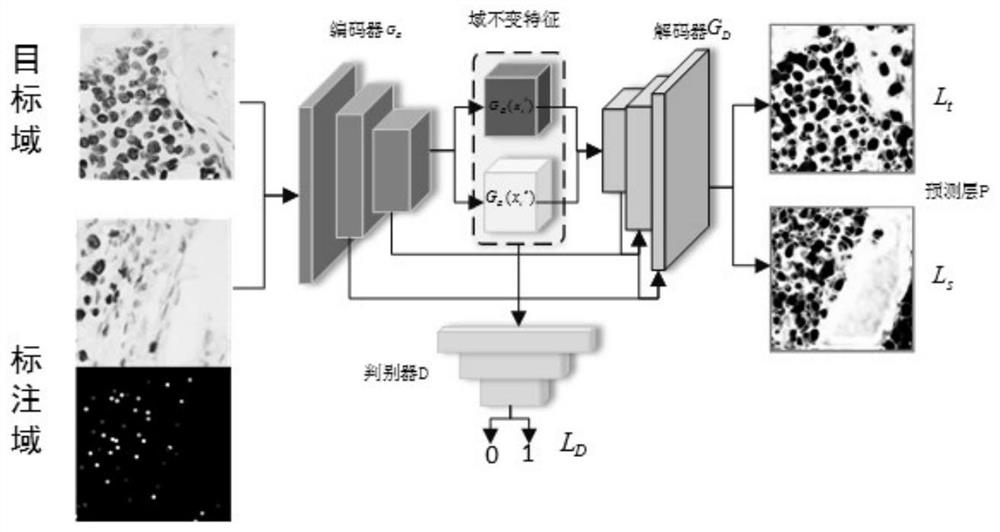
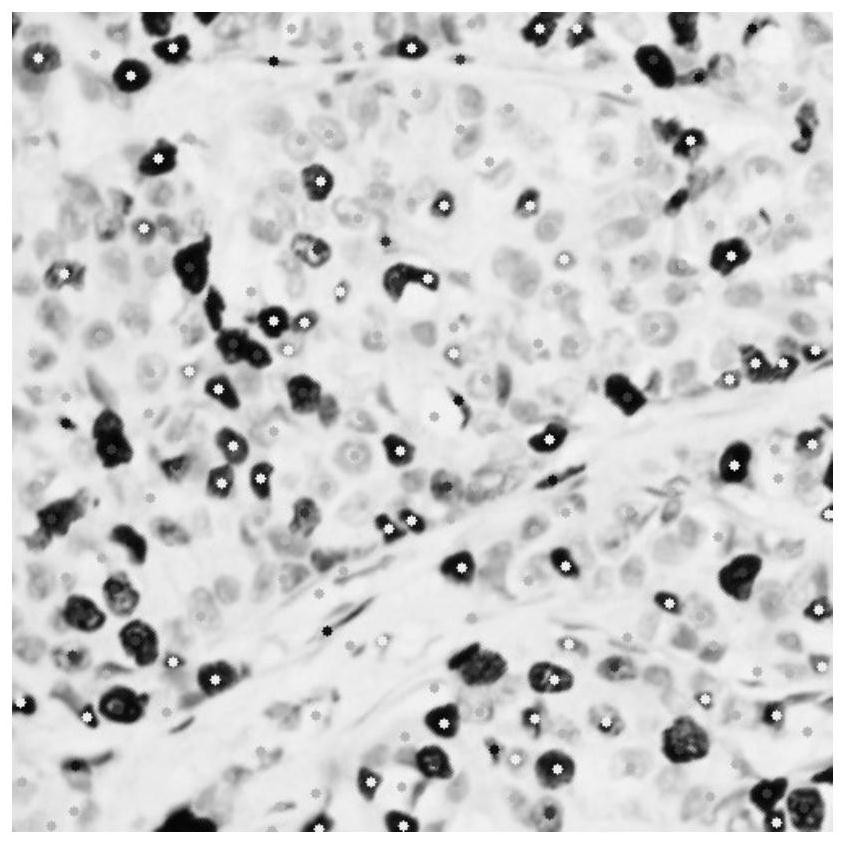
[0034] There are often hundreds of thousands of cells in pathological images. For immunohistochemical nuclear staining sections, pathologists need to count the tumor cells in the sections. Therefore, effective accurate positioning and classification of tumor cells is the assistant that pathologists currently desire. One of the tools. On the basis of only relying on point-level labeling, the cell key point positioning network effectively encodes the nucleus and its context information through the convolutional neural network, and then uses...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com