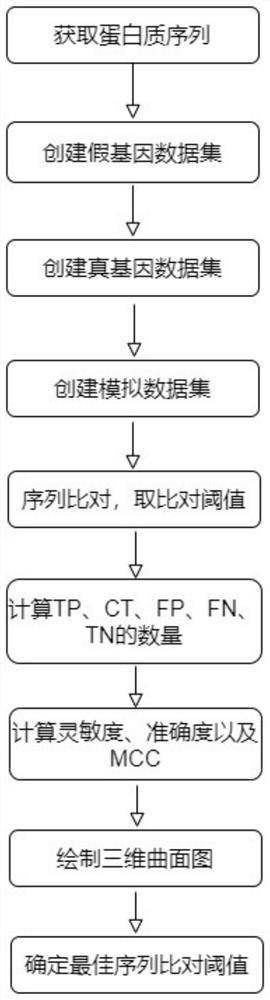
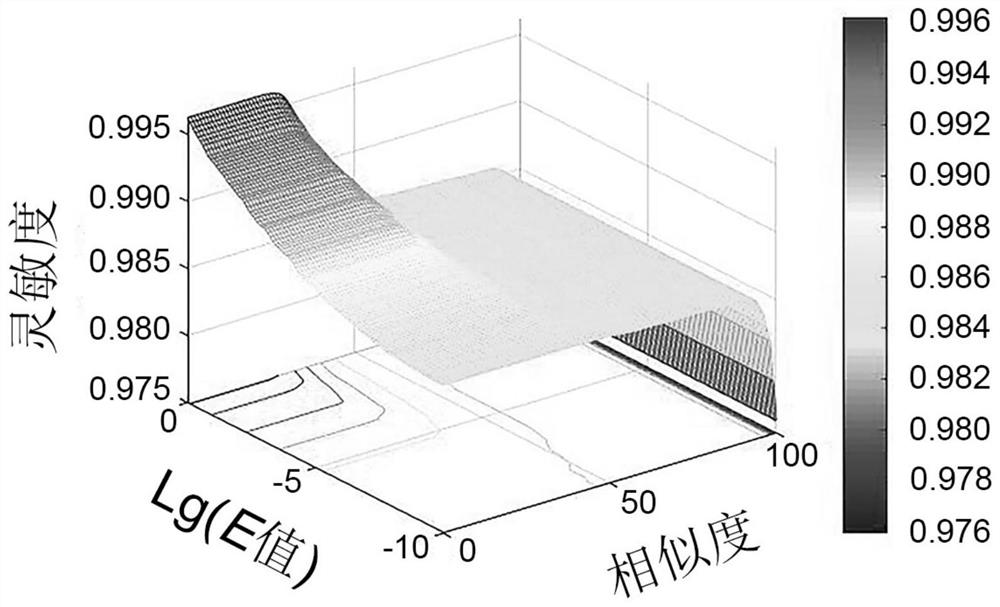
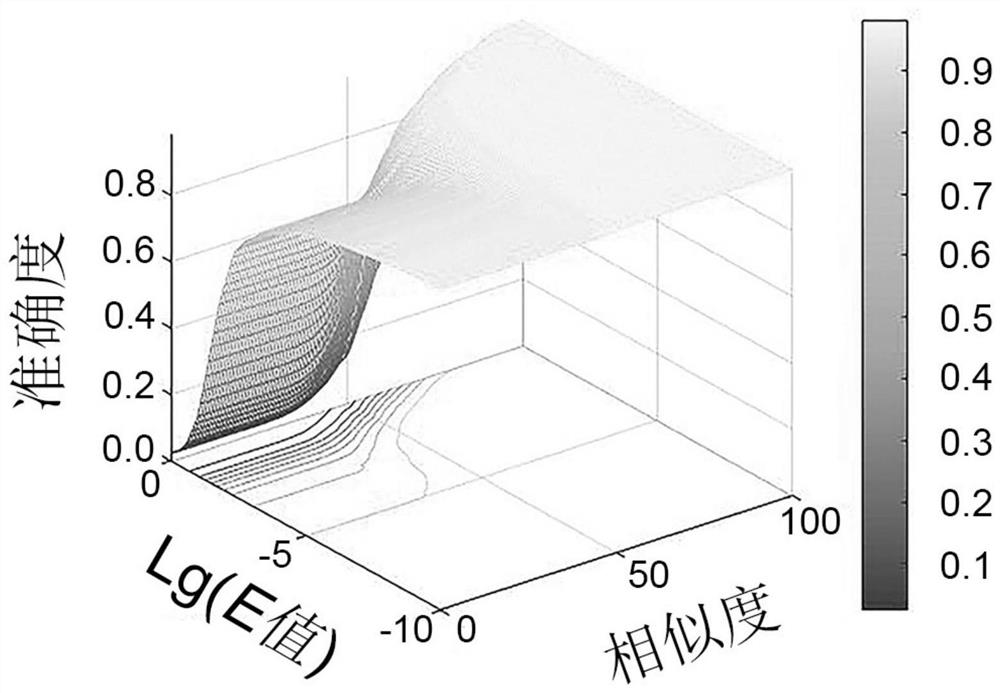
Method for determining optimal sequence alignment threshold for gene database
A technology for sequence alignment and gene data, which is applied in the biological field to achieve accurate alignment results and superior alignment performance.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0052] Taking the quorum sensing gene database as an example, the process of determining the optimal sequence alignment threshold (ie, similarity and E value) is described in detail below.
[0053] Step 1), in the Swiss-Prot protein sequence database of the UniProt protein database (https: / / www.uniprot.org / uniprot / ?query=reviewed:yes), download all protein sequences to the local, a total of 557134 protein sequences .
[0054] Step 2), from the 557,134 protein sequences obtained in step 1), remove the protein sequences (245 in total) that have been included in the quorum sensing gene database, and use the remaining 556,889 protein sequences as a false quorum sensing gene data set . For the protein sequences in the fake quorum sensing gene data set, "F" is marked after the sequence name, taking the sequence F4HRV8 as an example, marked as follows:
[0055]
[0056] Step 3), divide the protein sequences in the quorum sensing gene database into 11 subcategories according to t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com