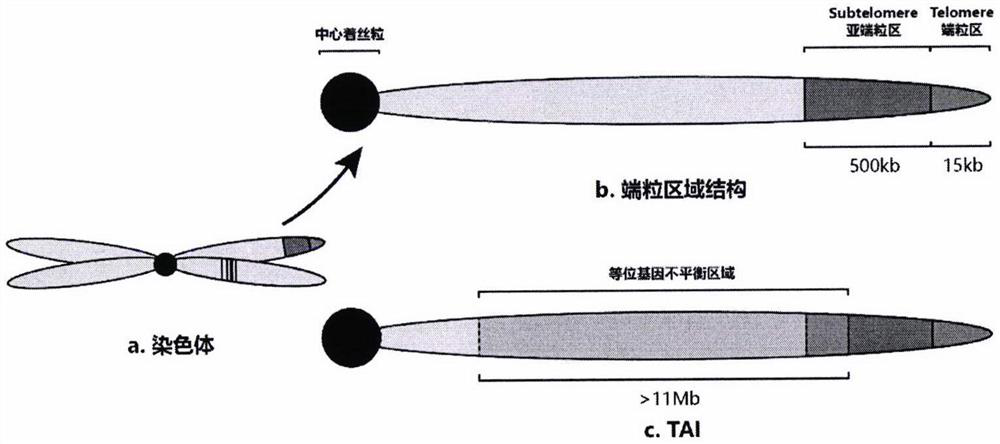
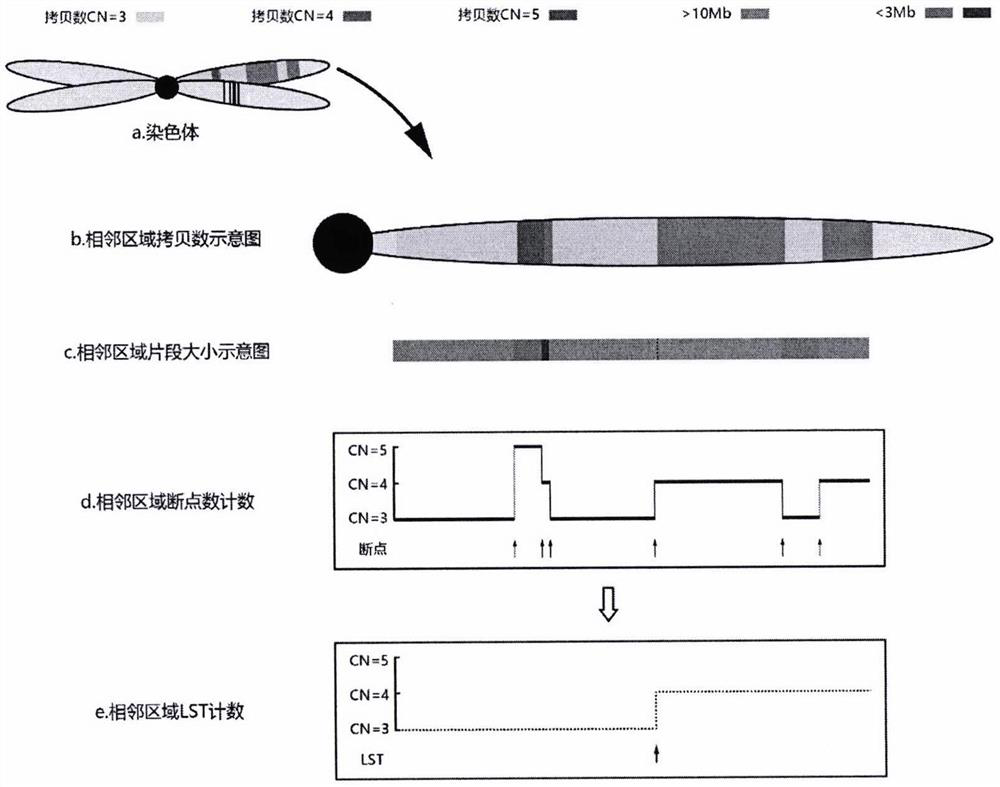
HRD score calculation method based on chip detection
A calculation method and chip detection technology, applied in biochemical equipment and methods, sequence analysis, microbial measurement/inspection, etc., can solve the problems of comprehensive scoring of homologous recombination defects and the inability to accurately judge the status of homologous recombination defects, etc. Achieve the effect of low cost, convenient and quick operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] The present invention provides such Figure 1-2 A method for calculating HRD scores based on chip detection is shown, and the specific operation steps are as follows:
[0034] 1): Take FFPE (Formalin-Fixed and Paraffin-Embedded, FFPE) samples from confirmed tumor patients to make H&E stained sections, observe the tumor cell content (TC%, Tumor Cell Percentage), and confirm that the tumor cell content is not less than 30%;
[0035] 2): Take the FFPE sample that has been screened for tumor cell content in step 1), and extract DNA (commercial FFPE sample DNA extraction kit can be used), the extraction concentration is required to be not less than 12ng / μL, and the extraction volume is not less than 6.6μL ;
[0036] 3): Using Affymetrix CNV FFPE Assay Kit processes the DNA solution extracted in step 2), after annealing (Anneal), gap filling (Gap Fill), first-stage polymerase chain reaction (1st PCR), second-stage polymerase chain reaction (2nd PCR), digestion (Hae III Di...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com