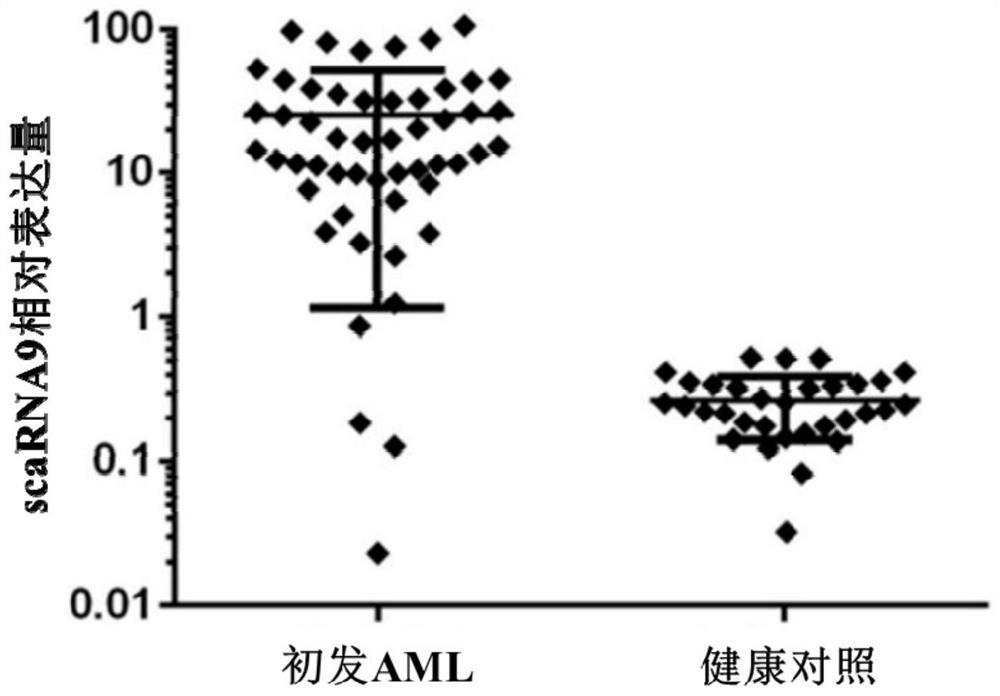
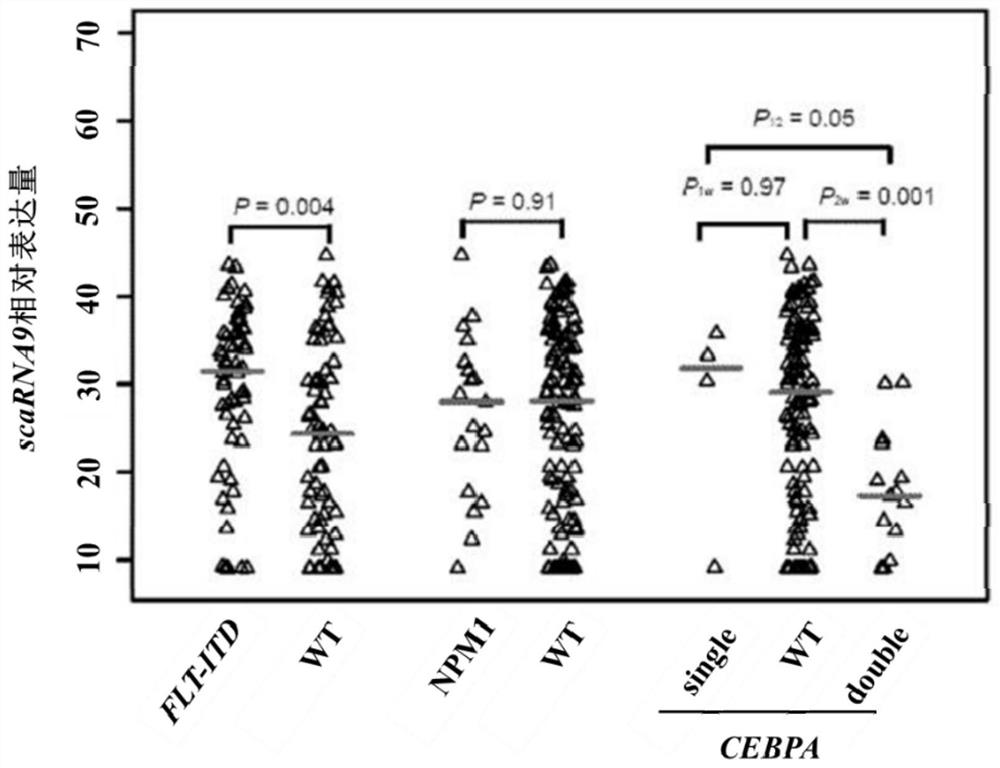
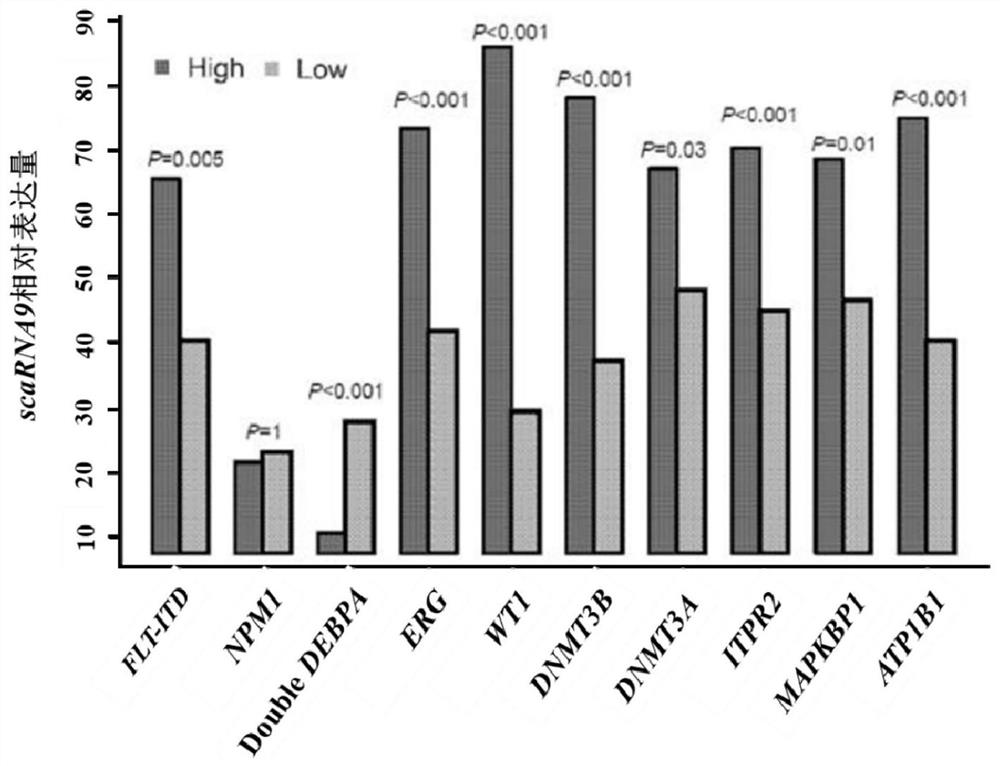
Application of scaRNA9 gene in early judgment of prognosis of acute myeloid leukemia
A technology for acute myeloid and leukemia, applied in the field of leukemia diagnosis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Example 1 extracts total RNA
[0040] Cell lysis: Add 1mL of QIAZOL reagent to the collected cell pellet, shake and mix well, and let it work at room temperature for more than 15 minutes to fully lyse it. If RNA cannot be extracted immediately after adding QIAZOL reagent, it can be stored at -20°C and can be used in a short time.
[0041] According to the chloroform:QIAZOL volume ratio of 1:4, add about 300 μL of chloroform, mix up and down for 1 minute, let stand at room temperature for 5-10 minutes, and centrifuge at low temperature: 4°C, 12600rpm for 10 minutes.
[0042] After centrifugation, the liquid in the EP tube is divided into three layers, the upper layer is the supernatant containing RNA, and the middle and lower layers are DNA and protein. Then, transfer the supernatant to a new EP tube, add an equal volume of isopropanol, mix up and down, let stand at low temperature for 10 minutes, and centrifuge: 4°C, 12600rpm for 15 minutes.
[0043] Discard the super...
Embodiment 2
[0046] Example 2 RNA reverse transcription
[0047] The reverse transcription system was prepared according to Table 1, and the total reaction volume was 25 μL (total RNA amount was 1 μg). The following operations were performed on ice:
[0048] Table 1 Preparation of reverse transcription system
[0049] components Reaction tube volume (μL) 10nM dNTPs 1.2 reverse transcriptase MMLV 1.3 MMLV RT 5*buffer 5.0 RNAase inhibitor 0.6 random primer 1.0 nuclease free water 15.9 RNA 1μg total 25
[0050] After preparing the above reaction components into 0.2mL PCR reaction tubes, put them into a PCR instrument for reverse transcription reaction, the program is 37°C, 2 hours; 4°C, forever. The product obtained after the reaction is cDNA, and the reaction product is taken out and stored at -20°C until use.
Embodiment 3
[0051] Embodiment 3 real-time fluorescent quantitative PCR detection
[0052] Prepare the RT-PCR mix according to Table 2.
[0053] In the reaction tube, prepare 2 times the volume of 2×SYBR Green I, Nuclease-free water, template cDNA and ROX II mixture, mix well and divide into two 0.2mL PCR reaction tubes, and then add the target gene scaRNA9 respectively The primer for mRNA (SEQ ID NO.1) was paired with SEQ ID NO: 2 and 3 or the primer for internal reference GAPDH (glyceraldehyde-3-phosphate dehydrogenase, glyceraldehyde-3-phosphate dehydrogenase), and mixed gently.
[0054] scaRNA9 upstream primer (SEQ ID NO.2): 5'-TGAGCATCAGAAGTCTTTCCAGTC-3';
[0055] scaRNA9 downstream primer (SEQ ID NO.3): 5'-CACCCTCAATCTCATTCATTCCTAC-3'.
[0056] Table 2 Preparation of RT-PCR mix
[0057] components Reaction tube volume (μL) 2*SYBR Green I 12.5 ROX II 0.5 10μM Primer (F+R) 1.2 cDNA 1.8 Nuclease-free water 9.0 total 25
[0058] Put...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com