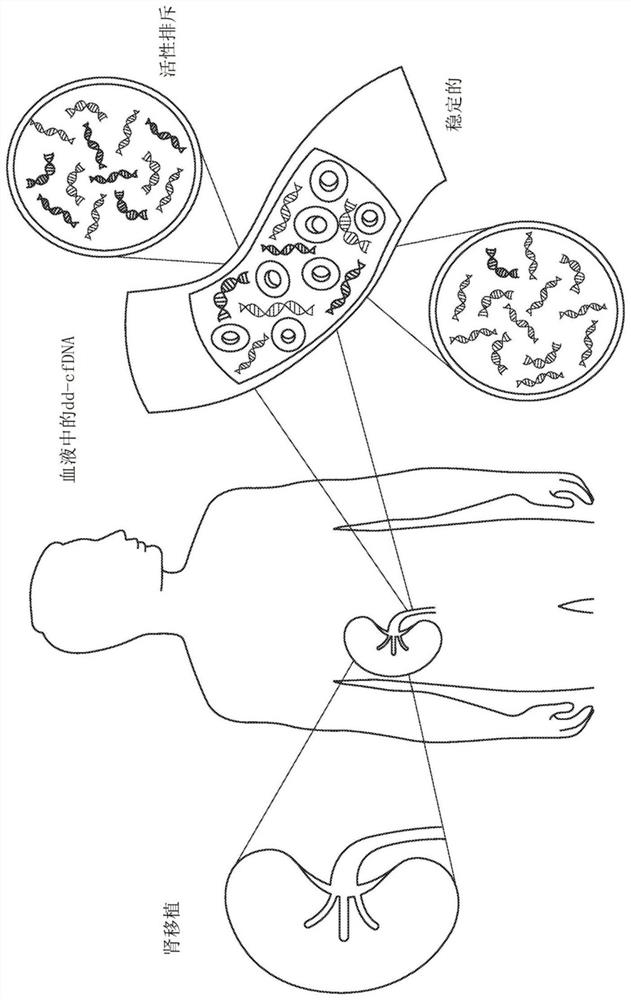
Methods for detection of donor-derived cell-free DNA
A source and cell technology, applied in the direction of biochemical equipment and methods, microbial measurement/testing, etc., can solve the problems of invasive costs, transplant damage, lack of specificity, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
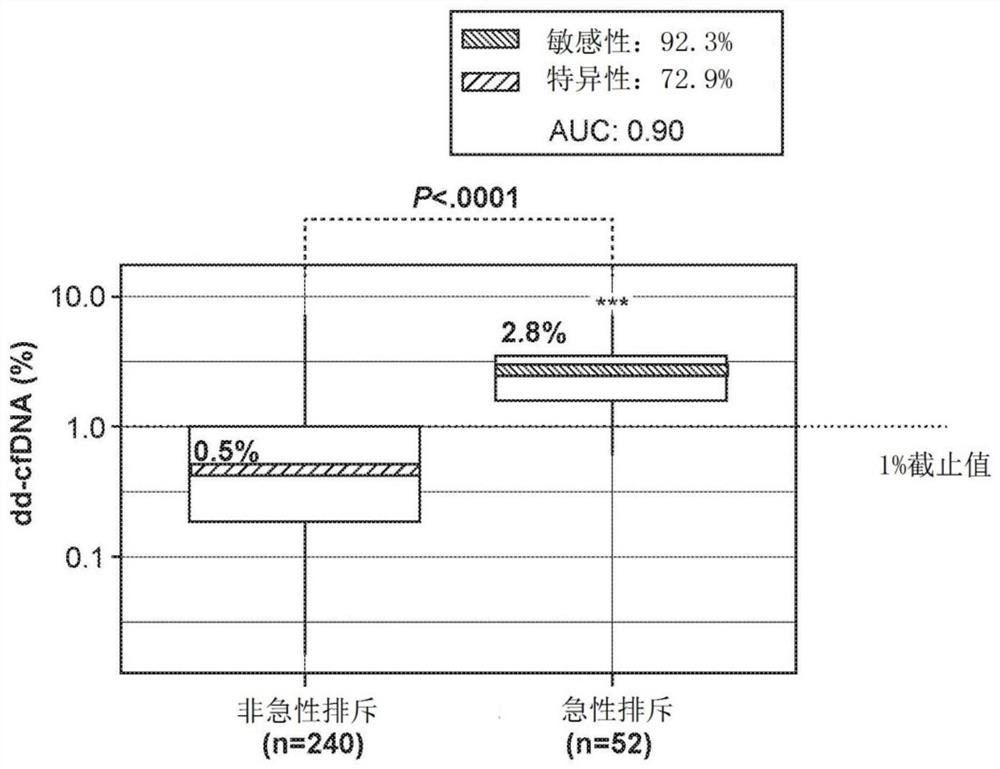
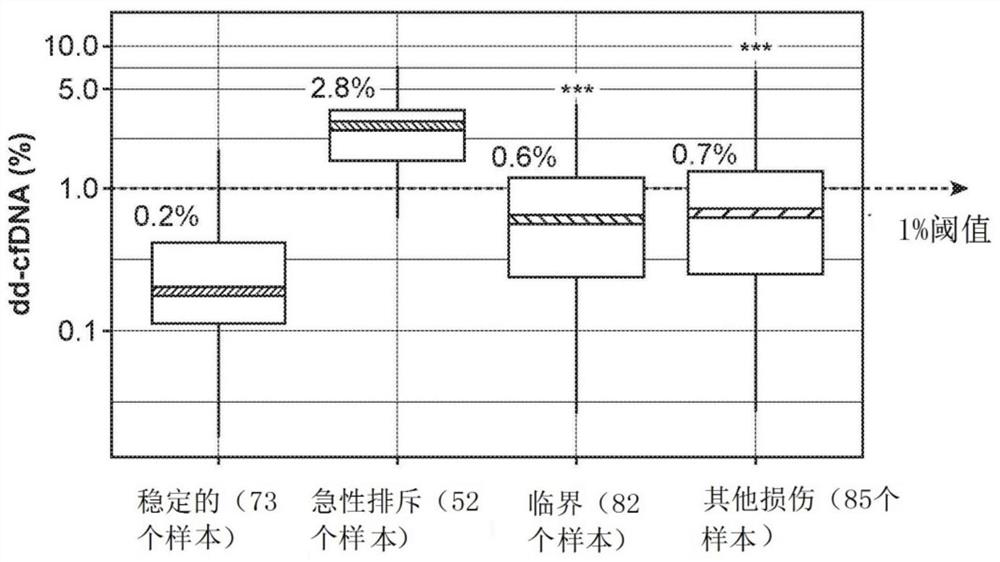
[0769] Blood samples from kidney transplant recipients were analyzed retrospectively (292 plasma samples from 187 unique patients, of which 8 samples were excluded) after assessment of the patients' graft status by biopsy. Biopsies were graded by the Banff classification of T-cell and antibody-mediated acute rejection (AR) or non-AR (borderline, stable, or other injury). The samples analyzed for biopsy included 52 acute rejection (AR) samples and 240 non-acute rejection (non-AR) samples, including those in borderline rejection, with other injuries or stable.
[0770] Circulating cell-free DNA was extracted from 2 mL of plasma per sample by Qiagen cfDNA kit. The amount of cfDNA was then quantified using a LapChip. Library preparation was accomplished using the standard protocol using the Natera Panorama Library Prep Kit, except that the library was amplified by 18 cycles of PCR (as opposed to the standard 9 cycles). The amplified library was then purified using Ampure beads (...
example 2
[0782] Example 2. Optimization of detection of kidney transplant injury by assessment of donor-derived cell-free DNA via large-scale multiplex PCR.
[0783] introduce
[0784] Precision medicine and personalized customization of immunosuppressive drug regimens could improve the current state of organ transplant management. Invasive biopsies should be avoided where possible considering patient experience, and graft damage is usually detected late. Although advances in immunosuppressive drugs, organ harvesting methods, and human leukocyte antigen typing have reduced the number of clinically and biopsy-proven acute rejection events, subclinical acute rejection of kidney transplantation remains a significant risk. Kidney transplant management is particularly challenging due to the redundancy of serum creatinine measurements, which makes immunosuppressant dosing and adjustment far from individualized, in addition to late detection of graft injury. Therefore, rapid and non-invasiv...
example 3
[0847] Example 3. Evaluation of donor-derived cell-free DNA by large-scale multiplex PCR and next-generation sequencing to validate the detection of kidney transplant injury.
[0848] introduce
[0849] Kidney transplantation is the best option for patients with end-stage renal disease. According to the United Network for OrganSharing, more than 19,000 kidneys were transplanted in the United States in 2016 (cen.acs.org), and approximately 200,000 patients survived a functional kidney transplant (NIH Medline plus). Although life-long immunosuppressive maintenance regimens are designed to optimize treatment outcomes, approximately 20-30% of patients experience overall kidney transplant failure within the first 5 years, and only 55% of transplanted kidneys survive to 10 years (cen.acs.org ). Therefore, early intervention strategies are urgently needed to avoid or minimize acute / subclinical rejection episodes, nephrotoxicity, and enable management and monitoring of comorbidities...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com