A biosensor based on luciferase complementation and its preparation method and application
A biosensor, luciferase technology, applied in the field of biochemistry, can solve the problems of deviation in detection results, long detection time, complicated operation, etc., and achieve the effects of low background noise, low cost and high detection sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0073] The present invention also provides a method for preparing the above-mentioned biosensor based on luciferase complementation, including:
[0074] Constructing the first expression vector containing the luciferase amino-terminal fragment gene, and constructing the second expression vector containing the luciferase carboxy-terminal fragment gene, wherein the 5' end of the luciferase amino-terminal fragment gene in the first expression vector, and one of the 3' ends of the luciferase carboxy-terminal fragment gene in the second expression vector is inserted into the G protein gene, and the other is inserted into the gene of the first marker;
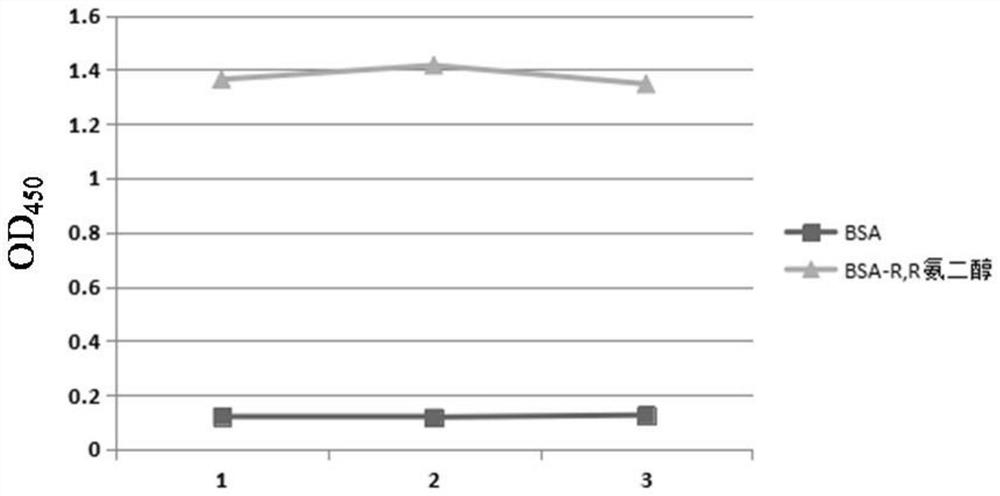
[0075] The first expression vector and the second expression vector are transformed, expressed and purified to obtain the luciferase amino-terminal fragment and the luciferase carboxy-terminal fragment, the amino-terminal of the luciferase amino-terminal fragment and the carboxyl group of the luciferase carboxyl-terminal fragment One...
Embodiment 1
[0093] A kind of preparation method of biosensor based on luciferase complementation
[0094] (1) Expression and purification of fusion protein
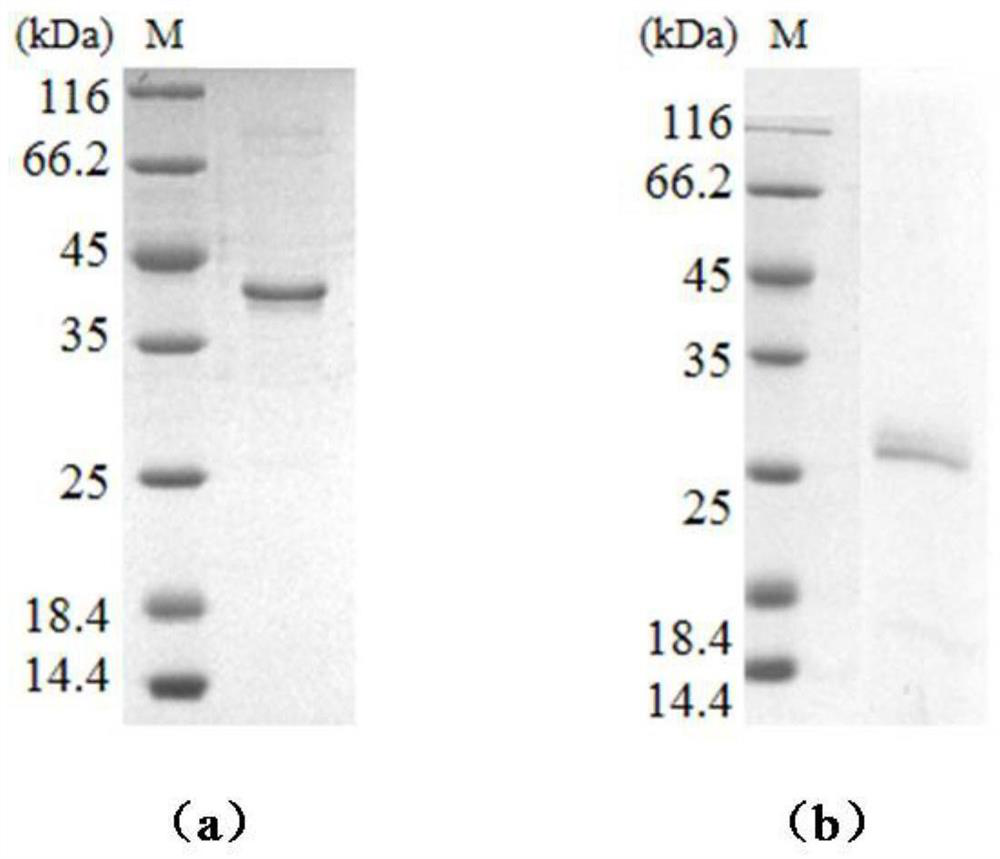
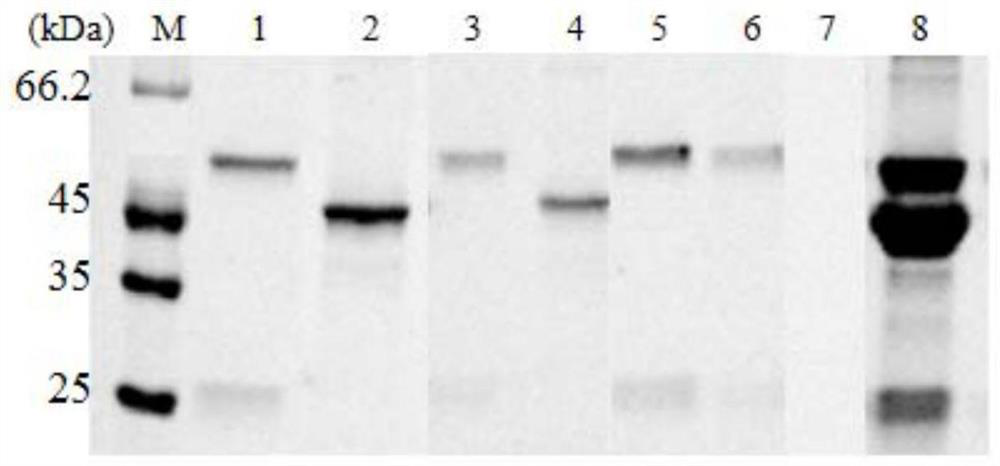
[0095] Gaussia luciferase (Gluc) was divided between the G93 and E94 sites to form two complementary N-terminal fragments and C-terminal fragments, and the coding sequences of the N-terminal fragments and C-terminal fragments were synthesized. Pass the 5' end of the coding sequence of the N-terminal fragment through (GGGGS) 2 The coding sequence of the G protein is connected to the coding sequence of the His tag; the 3' end of the coding sequence of the C-terminal fragment is passed (GGGGS) 2 The coding sequence of the monomeric streptavidin (mSA) and the coding sequence of the His tag were connected and inserted into the pET21a expression vector respectively.
[0096] The above pET21a expression vector was transformed into competent cells BL21 after heat treatment at 42°C for 45s, revived by culturing at 37°C, spread on LB plates ...
Embodiment 2
[0104] A kind of preparation method of biosensor based on luciferase complementation
[0105] Two complementary N-terminal fragments and C-terminal fragments formed by dividing Renilla luciferase (Rluc) between L110 and P111 sites were synthesized into coding sequences of the N-terminal fragment and the C-terminal fragment. Pass the 5' end of the coding sequence of the N-terminal fragment through (GGGGS) 3 The coding sequence of the monomeric streptavidin (mSA) was joined to the coding sequence; the 3' end of the coding sequence of the C-terminal fragment was passed (GGGGS) 3 The coding sequence of G protein is connected with the coding sequence of G protein. The 3' end of the above gene sequence was connected to the gene encoding the His tag, and inserted into the pET21a expression vector respectively. According to the same method as in Example 1, luciferase amino-terminal fragment (mSA-NRluc) and luciferase carboxyl-terminal fragment (CRluc-potein G) were prepared, and the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com