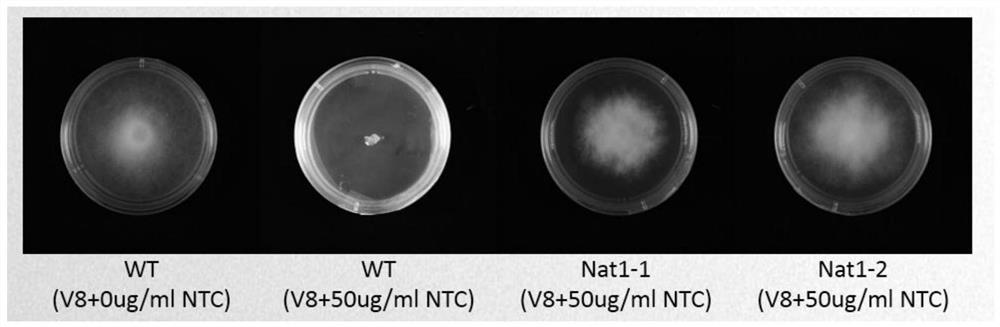
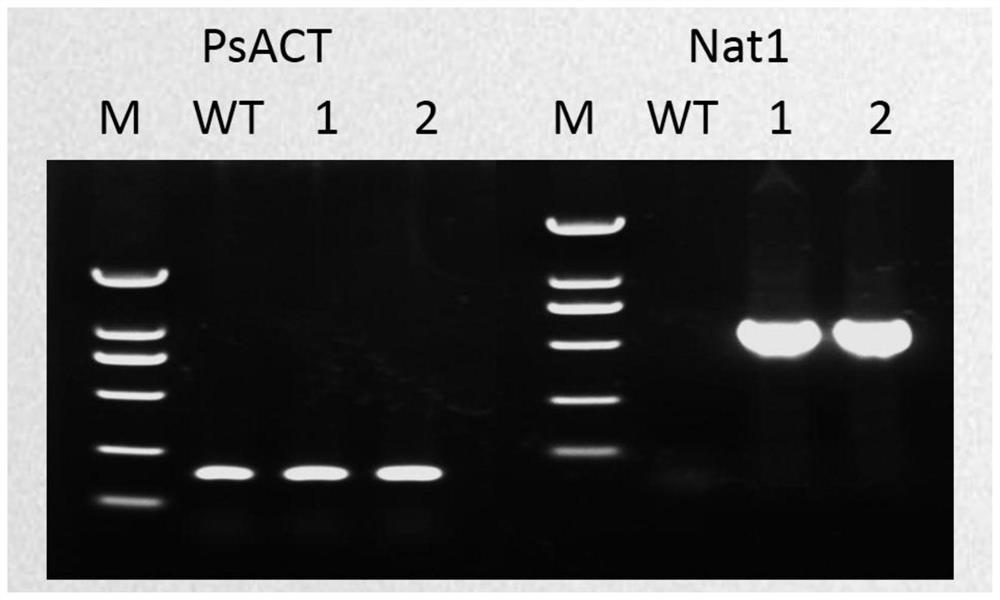
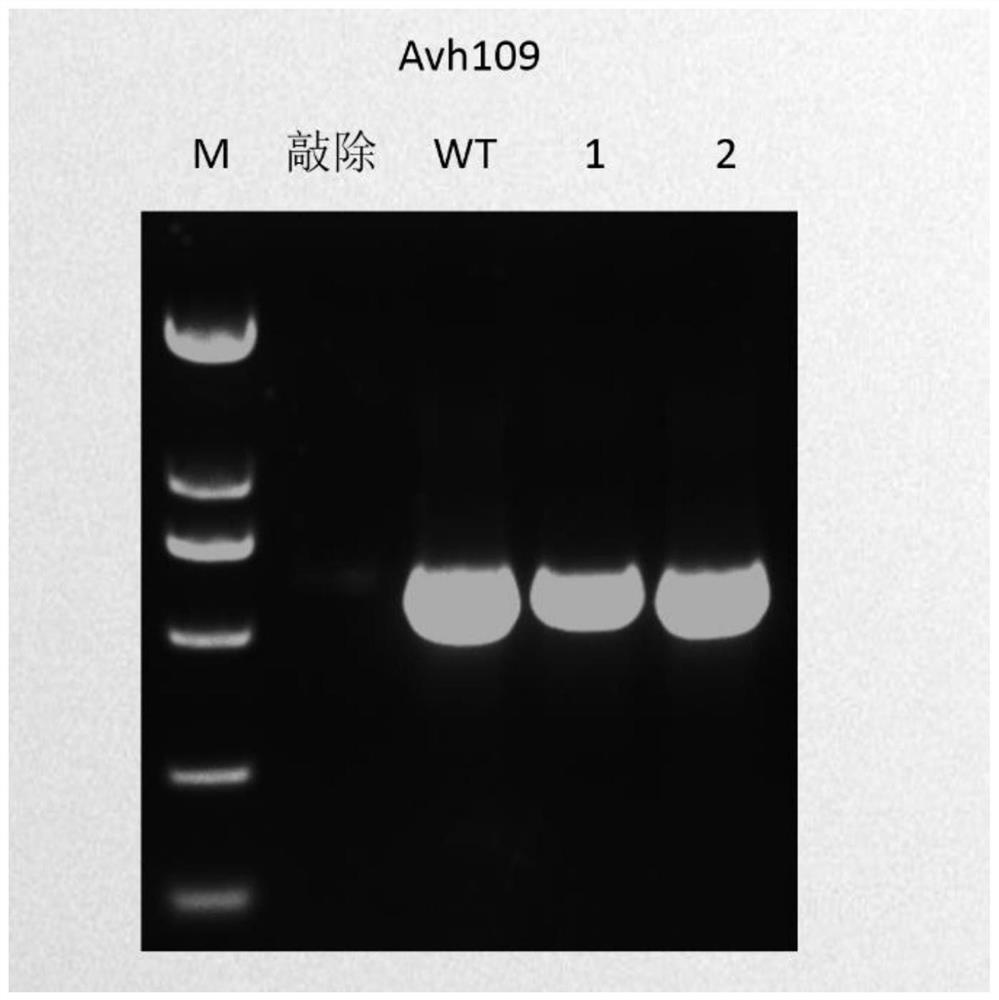
Application of Nat1 gene as selection marker in genetic transformation of oomycetes
A technology of genetic transformation and screening markers, applied in the field of genetic engineering, can solve the problem of no screening markers for oomycete genes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Embodiment 1: Completion and screening of Phytophthora soybean Avh109 gene
[0045] 1. Clone the Nat1 gene and the target gene (Avh109 gene) fragment:
[0046] The following primers Nat1-F and Nat1-R were used to clone the Nat1 gene fragment, and the nucleotide sequence of the cloned Nat1 gene fragment was shown in SEQ ID NO.1.
[0047] Nat1-F: 5'-ACACAAGGGCCCCGTTTCGCATGGGTACCACTCTTGACG-3'; (SEQ ID NO.2)
[0048] Nat1-R: 5'-TTCGAACCCCAGAGTCCCGCTTAGGGGCAGGGCATGCT-3'; (SEQ ID NO.3)
[0049] Search the PsAvh109 gene from the NCBI database (http: / / www.ncbi.nlm.nih.gov / ), download its CDS and genome sequence, design primers G Avh109-F and G Avh109-R, and clone the Avh109 gene fragment, which The sequence is shown in SEQ ID NO.4.
[0050] G Avh109-F: 5'-CCatcgatATGCGTCTCCAGTATGCCG-3'; (SEQ ID NO.5)
[0051] G Avh109-R: 5'-GgaattcATCAGCGGTTTGTCGCC-3'; (SEQ ID NO.6)
[0052] 2. Construct the target gene (Avh109 gene) and Nat1 gene in the complementation vector pTOR:
[00...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com