Construction method of disease animal model and fusion protein
A technology of fusion protein and purpose, applied in the field of genetic engineering, can solve problems such as not very effective, and achieve the effect of improving editing efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0054] The present invention will be described in further detail in conjunction with the following specific examples and accompanying drawings, and the protection content of the present invention is not limited to the following examples. Without departing from the spirit and scope of the inventive concept, changes and advantages conceivable by those skilled in the art are all included in the present invention, and the appended claims are the protection scope. The process, conditions, reagents, experimental methods, etc. for implementing the present invention are general knowledge and common knowledge in the art except for the content specifically mentioned below, and the present invention has no special limitation content. As described in Sambrook et al., Molecular Cloning, A Laboratory Manual (New York: Cold Spring Harbor Laboratory Press, 1989), or according to the manufacturer's suggested conditions.
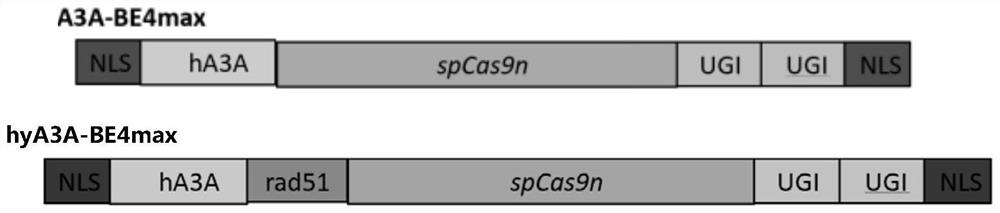
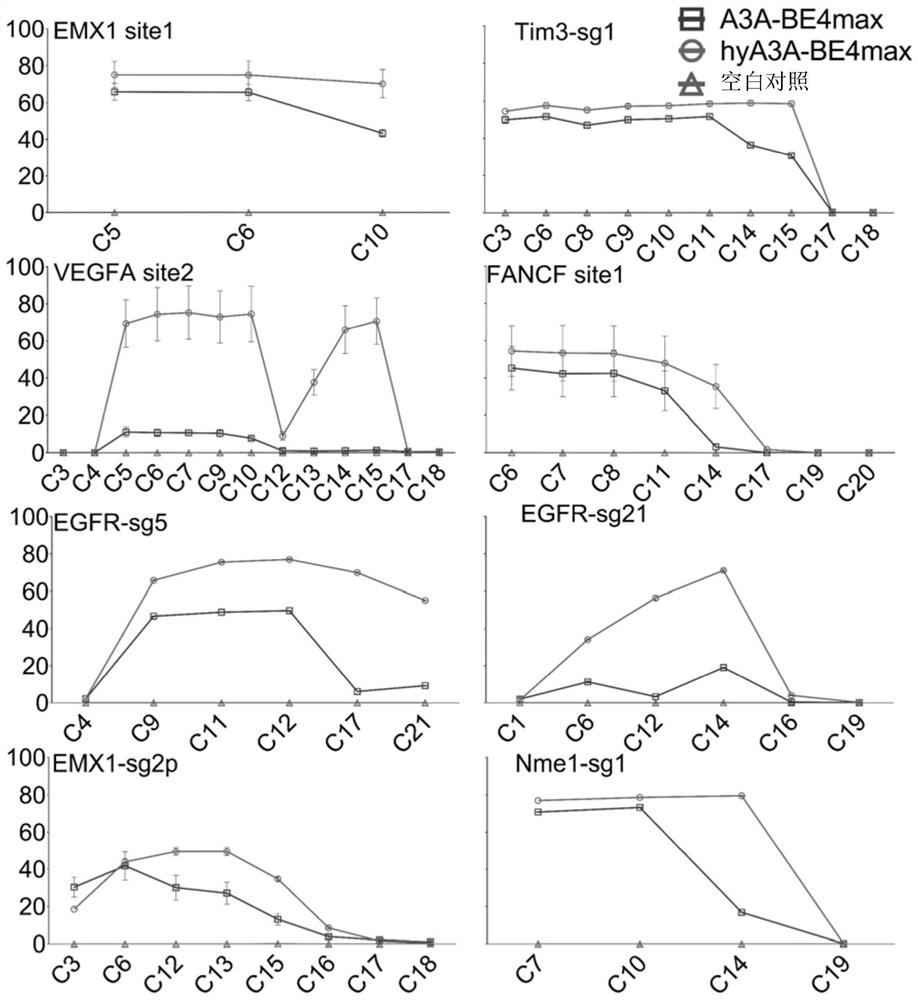
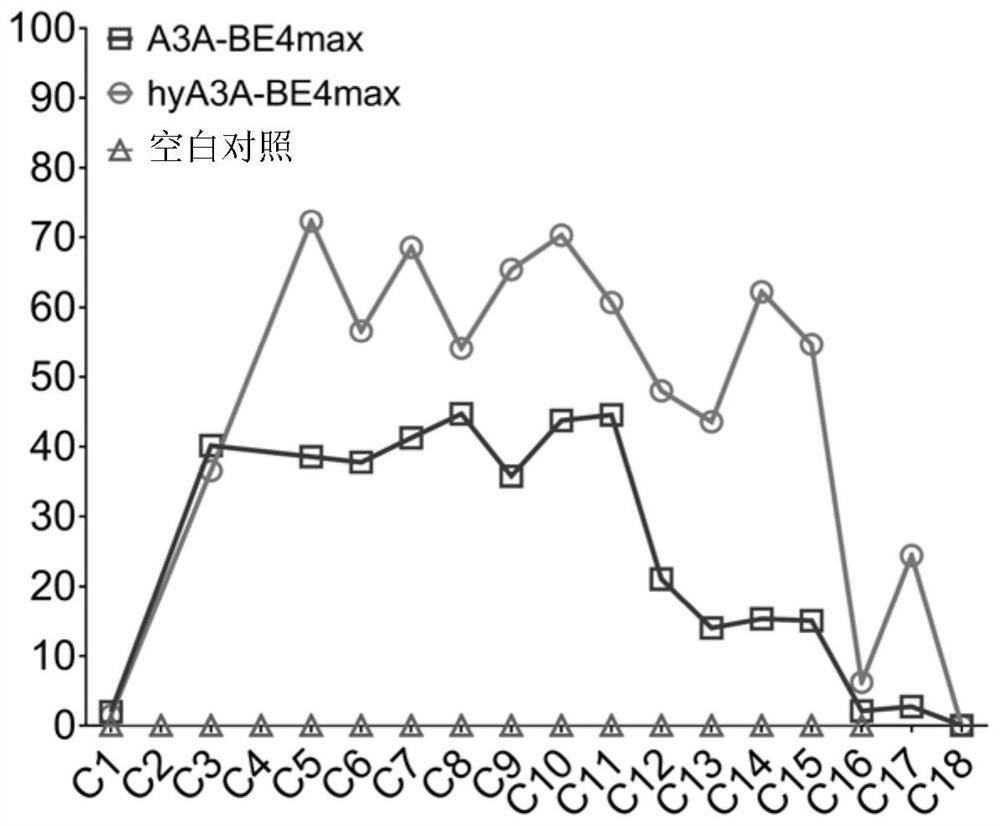
[0055] 1. Working characteristics of fusion protein hyA3A-BE4max
[005...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com