Mouse model with miRNA-125a knocked in based on CRISPR/Cas9 technology and construction method
A mouse and technology technology, applied in the field of genetic engineering, can solve problems such as the lack of C57BL/6J mouse construction method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] Example 1 Design sgRNA
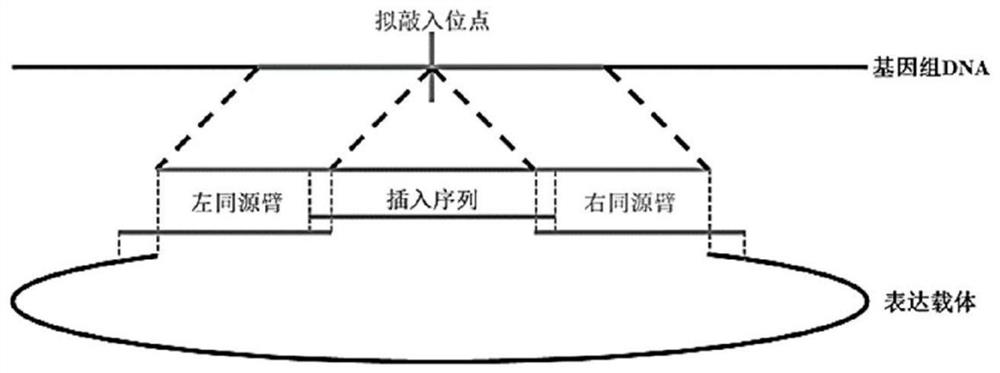
[0056] Compare the human and mouse sequences in this region in ensemble, and select the sequence difference site as the region to be knocked in (design sgRNA in this region) to ensure that the splicing information is not destroyed. Available sequences in the mouse genome were analyzed using the website (http: / / crispr.mit.edu). We selected one sgRNA (SEQ ID NO:1) as the site to be knocked in, as follows:
[0057] The sequence of SEQ ID NO:1 is (sgRNA1): CTCACAGGTTAAAGGGTCTC(5'→3')
[0058] According to the characteristics of the sticky end produced by the restriction endonuclease digestion of the vector, in order to make the sgRNA fragment connect to the linearized vector exactly, add GTTT before the 5' end of the forward strand, and add AAAC before the 5' end of the reverse strand. . The target sequences with adapters and their complementary sequences designed for the above sgRNAs are as follows:
[0059] The sequence of SEQ ID NO:2 is (sgRN...
Embodiment 2
[0061] Example 2: Construction of expression vectors
[0062] a) Plasmid linearization
[0063] Digest the lentiCRISPR V2 vector in the system shown in the table below:
[0064] Table 1 enzyme digestion system
[0065]
[0066] Digest overnight at 37°C, add loading buffer to a final concentration of not less than 1×, purify the digested product by 0.8% agarose gel electrophoresis, and recover the linear DNA band by cutting the gel. The steps for gel recovery are as follows (commercialized by Omega Corporation) Reagent test kit):
[0067] 1. Cut out the target DNA band in the agarose gel, add an equal volume of gel solution GSB, and put it in a metal bath at 65°C for 5-10 minutes to completely dissolve it.
[0068] 2. Add all the glue solution to the spin column in batches and let it stand for 1min, centrifuge at 12000rpm for 1min, and discard the effluent.
[0069] 3. Add 300 μL GSB to wash the spin column, centrifuge at 12000 rpm for 1 min, and discard the effluent.
...
Embodiment 3
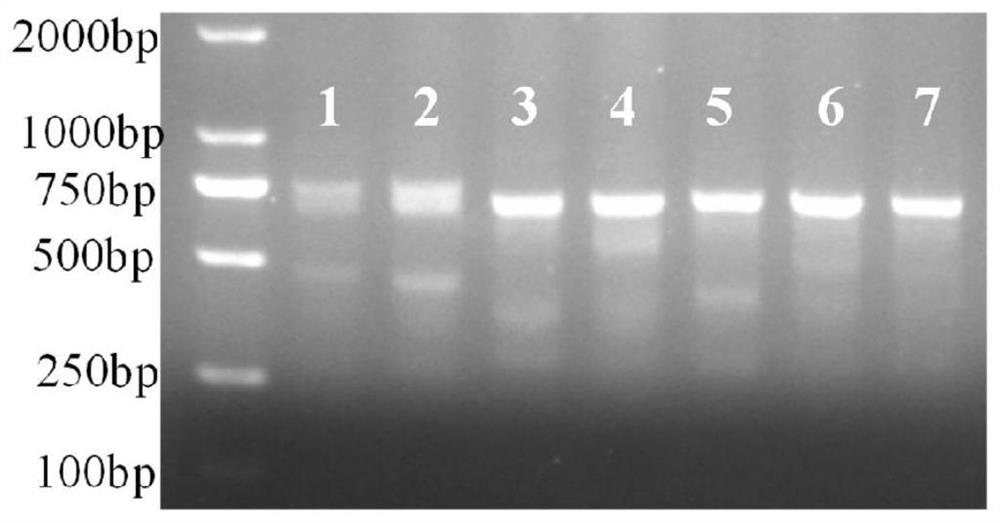
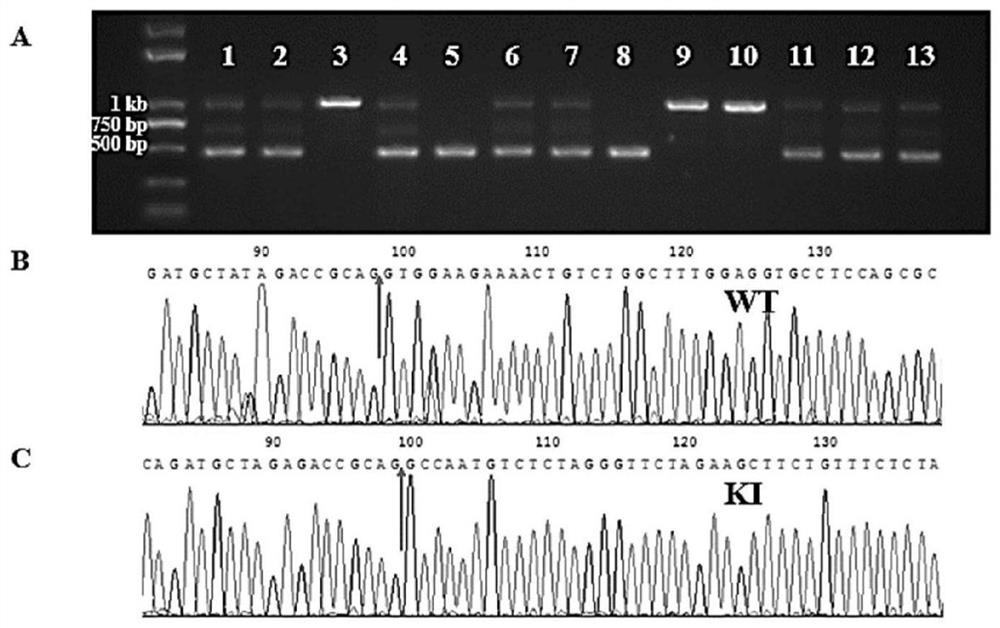
[0106] Example 3: Verification of sgRNA activity
[0107] 1) Routinely culture mouse liver AML12 cells, spread 12-well plates, and transfect the constructed LentiCRISPR V2 vector when the confluence is about 80%, transfect 1.5 μg of plasmid per well.
[0108] 2) Prepare a complete medium containing 2 μg / mL puromycin (puro). After 24 hours of transfection, change to the selection medium containing puro, and continue to culture normally for 48 hours; at this time, it can be observed that the cells in the control wells without transfection plasmid gradually die, and continue to culture with the normal complete medium until the cells are basically confluent. The genomic DNA of the cells can be extracted, and the target fragment containing the sgRNA target region is amplified by PCR, and the PCR primers are designed as F: 5'-GATAGCGAAGTGGTGGTGGT-3' (SEQ ID NO: 4), R: 5'-TTCTGAAGGAGGAGGGGATT-3' ( SEQ ID NO:5), the full length is 735bp. The PCR amplification system is as follows: ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com