Application of biomarker for predicting sensitivity of tumor patient to specific anti-tumor drug
A technology of anti-tumor drugs and biomarkers, which can be used in compound screening, biological material analysis, biochemical equipment and methods, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0093] Example 1, the common mechanism of action of cisplatin and all PARP inhibitors
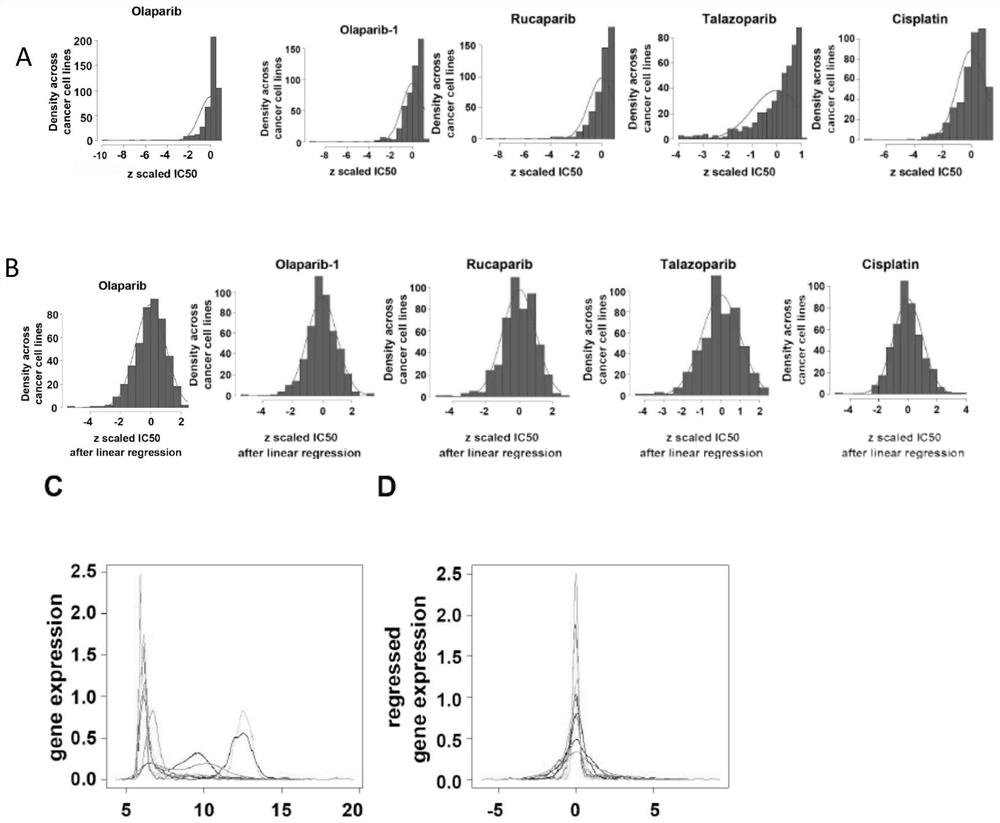
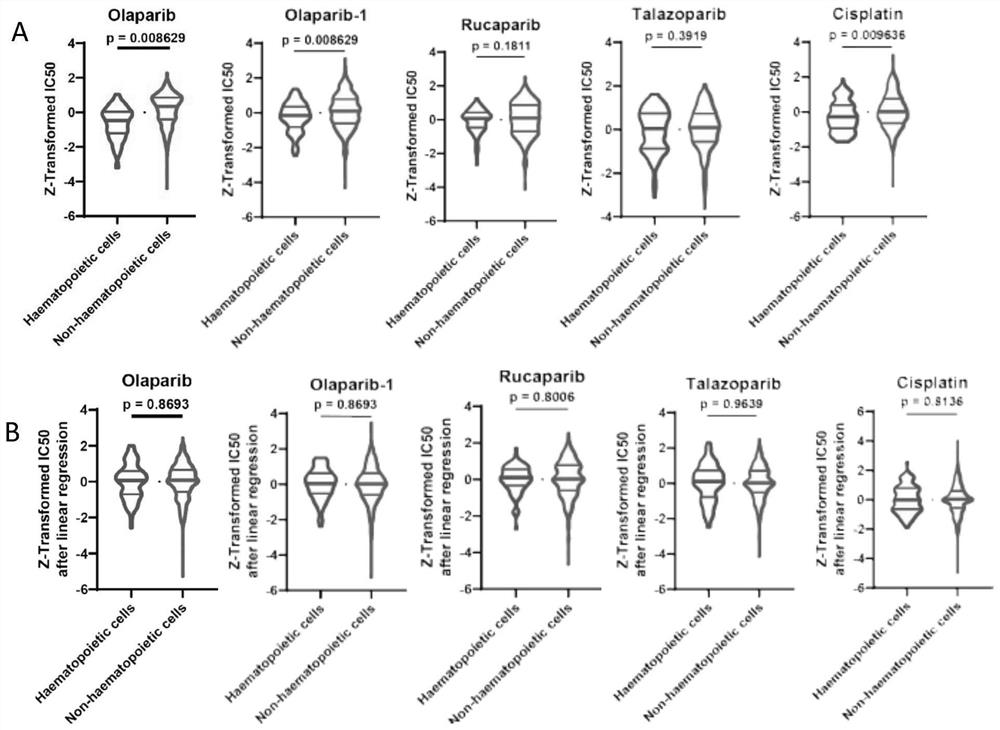
[0094] To study the different mechanisms of action of DNA damage therapeutics, big data analysis was performed. Drug sensitivity data (ln(IC50)) of human cancer cell lines from Genomics of Cancer Drug Sensitivity (GDSC), cell line RNA-seq data from Cancer Cell Line Encyclopedia (CCLE), RNA-seq data from RNA-seq data Only coding genes were selected. Unrecognized genes and genes whose expression was zero in all samples were excluded. For genes with multiple records, we calculated the sum of the expression levels of its multiple records as its expression level for subsequent Weighted correlation network analysis (WGCNA) and Pearson correlation analysis. Before downstream analysis, the RNAseq data of the remaining genes were VST transformed by the variance stable transformation (VST) function of DESeq2. Use based on tissue source and cancer subtype annotation (" ccle_primary_site", "ccle_prima...
Embodiment 2
[0101] Example 2: Correlation between ribosome biogenesis and cellular sensitivity to PARP inhibitors / cisplatin
[0102] To obtain biological pathways predictive of PARP inhibitor and cisplatin sensitivity, GO enrichment analysis was performed on genes in WGCNA-derived signature modules for these drugs. Signed co-expression networks were constructed using preprocessed coding gene expression levels by the WGCNA R package. Pearson correlation coefficients were calculated for each gene module between intrinsic genes and drug susceptibility (ln(IC50)) after pretreatment. The co-expressed gene modules most negatively correlated with IC50 constitute the drug's "signature". Genes in these WGCNA-derived drug signature modules were subjected to GO enrichment analysis by Gene Ontology analysis and visualization tools (Gorilla). Defined biological processes, molecular functions and cellular components GO terms included rRNA processing (p=4.48E -4 ), RNA binding (p=2.76E -5 ) and nucl...
Embodiment 3
[0134] Example 3: Prediction of cell line response to cisplatin and PARP inhibitors by associated gene sets
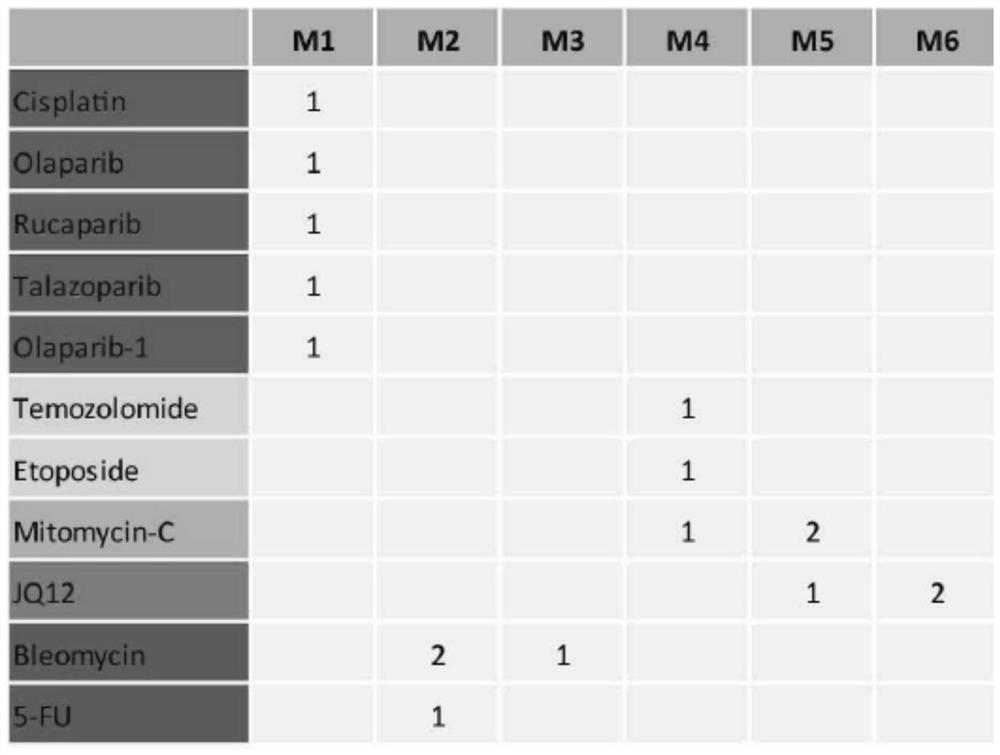
[0135] Based on the analysis of studies in the above examples, the ribosome synthesis pathway is a potential predictor of response to PARP inhibitors / cisplatin. Therefore, based on this drug mechanism, we developed a new strategy to select representative genes as biomarkers of cellular sensitivity to PARP inhibitors / cisplatin. Our hypothesis is that a subnetwork of drug targets involved in ribosomal synthesis has the potential to predict drug sensitivity. Drug signature modules may represent biological processes targeted by drug therapy, and genes negatively associated with sensitivity to these drugs may represent drug target genes. Therefore, we obtained multiple PARP inhibitor and cisplatin drug signature module genes and their respective genes negatively correlated with sensitivity (FDR<0.05); obtained 21 candidate genes (MYBBP1A, UBE2G1, EIF4A1, NUP88, GEMIN4 , P...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com