Spinal muscular atrophy detection method,
A spinal muscular atrophy and detection method technology, applied in the field of genetic detection, can solve the problems of insufficient comprehensiveness and accuracy of detection results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
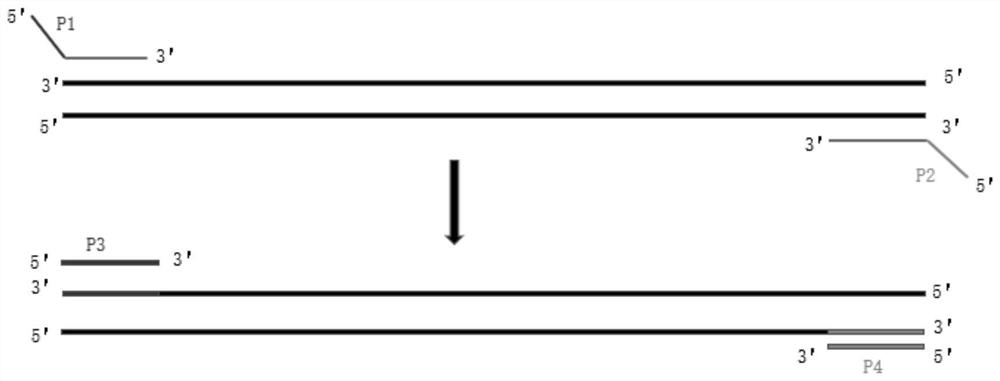
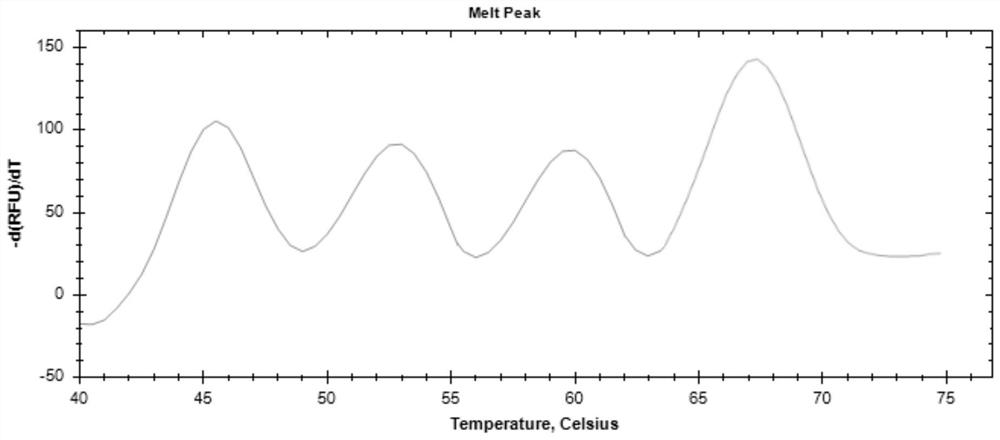
[0043] Optionally, the 5' ends of the nucleotide sequences four to seven are respectively connected to the FAM fluorescent group, and the 3' ends are respectively connected to the BHQ1 quenching group; the 5' ends of the nucleotide sequences eight to eleven are respectively The VIC fluorescent group is connected, and the 3' ends are respectively connected to the BHQ1 quenching group; the 5' ends of the nucleotide sequence twelve to fourteen are respectively connected to the ROX fluorescent group, and the 3' ends are respectively connected to the BHQ2 quenching group; The 5' ends of the 15th to 17th nucleotide sequences are respectively connected with a CY5 fluorescent group, and the 3' ends are respectively connected with a BHQ3 quenching group. The -39P detects the c.-39A>G site. The -7A5P detects c.-7_9del and c.5C>G sites. The 22P detects the c.22dupA site. The 40A43P detects c.40G>T and c.43C>T sites. The 56P detects the c.56delT site. The 84P detects c.84C>T. The 326...
Embodiment 2
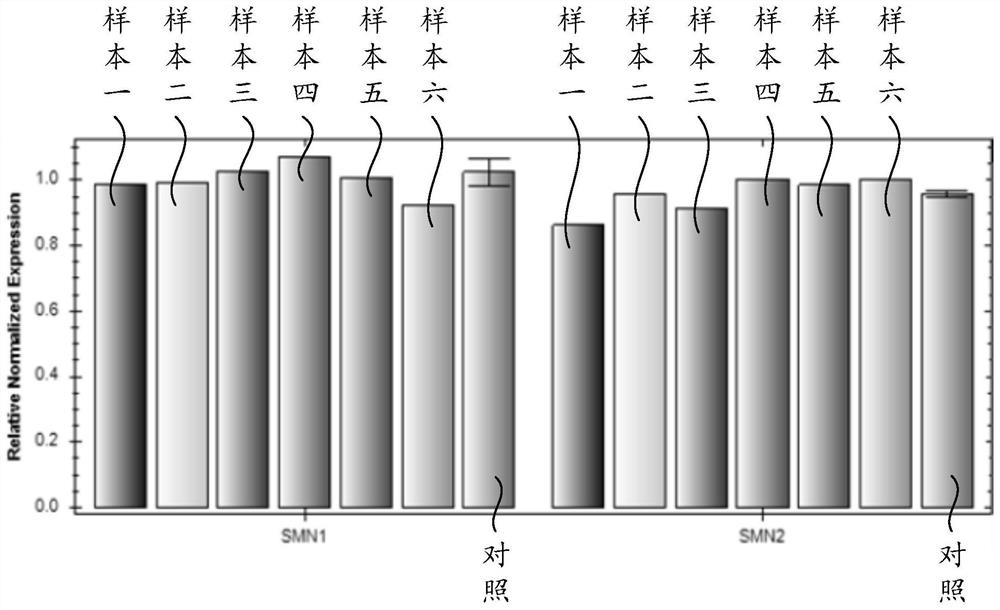
[0071] Using the detection method of the present invention, 150 cases of clinical samples were detected, and the detection results were compared with MLPA and Sanger sequencing methods. See Table 7 for test results.
[0072] Table 7 Test results of clinical samples
[0073]
[0074]
[0075]
[0076]
[0077]
[0078] The data in Table 7 shows that the detection result of the detection method of the present invention is completely consistent with the detection result of the prior art (MLPA and Sanger sequencing method), indicating that the accuracy of the detection method of the present invention is 100%, and the reliability of the detection method of the present invention is 100%. It is very high and can be used for clinical testing.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com