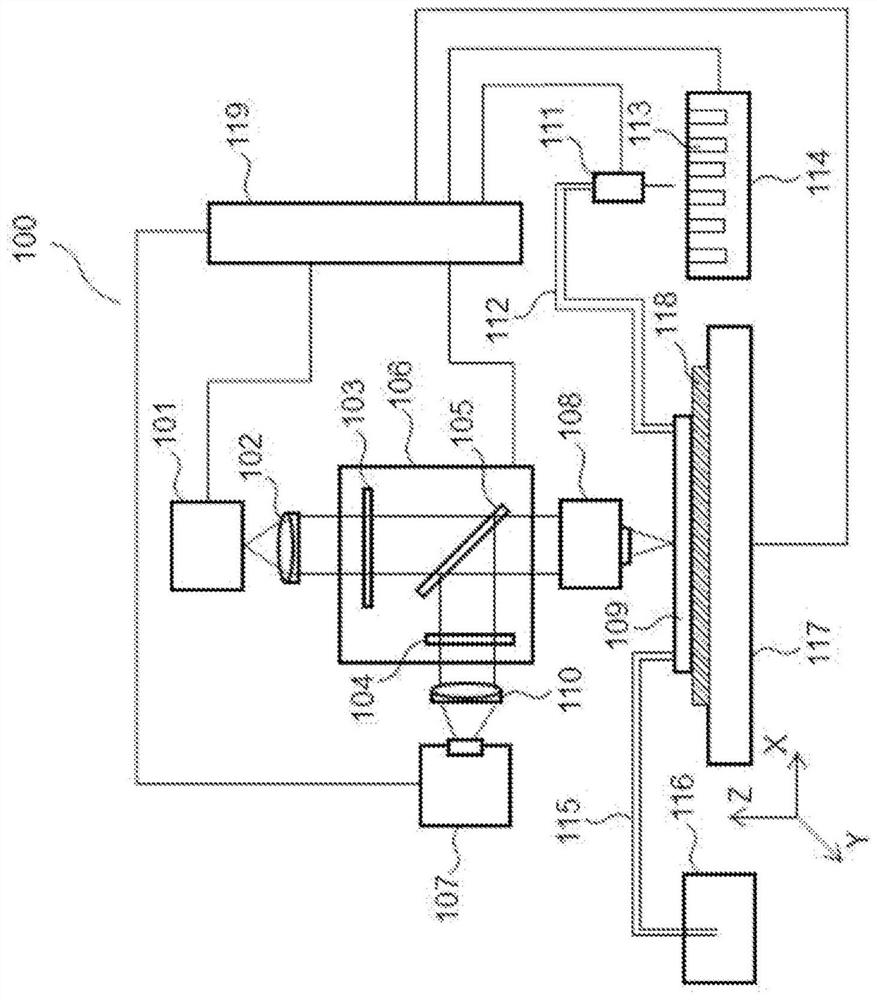
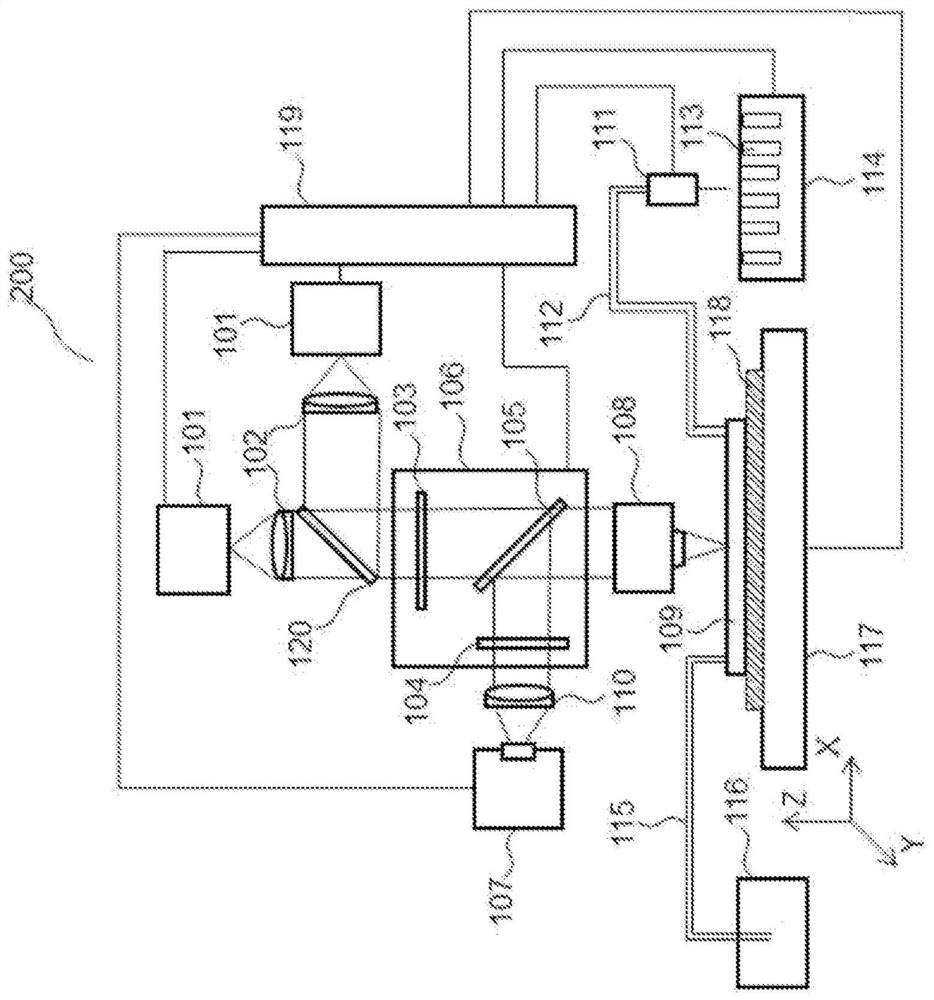
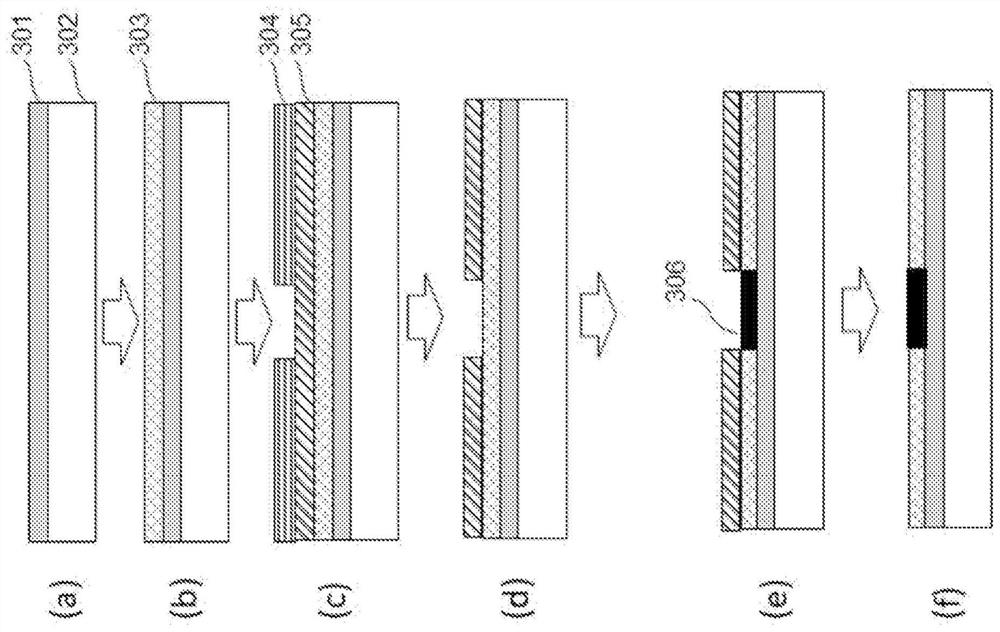
Substrate for nucleic acid analysis, flow cell for nucleic acid analysis, and image analysis method
A technology of nucleic acid analysis and analysis method, which is applied in the field of spot part configuration, can solve problems such as difficult identification of the position of nucleic acid attached spots, and achieve the effect of improving accuracy and processing speed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] use Figure 7 An example of a substrate for nucleic acid analysis including a patterned spot portion and a random spot portion on the surface of the substrate will be described.
[0059] Figure 7 It is an enlarged view of a part on the substrate. On the substrate, there are a patterned spot portion 701 , which is a region where nucleic acid attachment spots are regularly arranged, and a random spot portion 702 , which is a region where nucleic acid adheres irregularly. exist Image 6 In -A, the part where the circular parts are arranged represents the pattern-shaped spot part 701, and the circular parts represent the adhesion spots to which the sample adheres. Furthermore, the triangular part is a random spot part 702 . Each spot part has a nucleic acid-attached region formed of an amino group-containing coating film, and the surface of the non-nucleic acid-attached region is coated with hydrophobic HMDS. In the pattern-like spots, nucleic acid adheres to the arra...
Embodiment 2
[0066] An example of an image acquisition and alignment method using a nucleic acid analysis substrate having a patterned spot portion and a random spot portion will be described.
[0067] A nucleic acid sample to be analyzed is immobilized on a patterned spot portion and a random spot portion of a substrate disposed on the flow cell. Then, nucleotides with fluorescent substances are introduced by extension reaction, and four kinds of fluorescent images corresponding to four kinds of DNA bases are photographed and obtained. In each cycle of one base extension, four kinds of fluorescent images were observed as bright spots per field of view. exist Figure 9 , four examples of fluorescence images are shown. White circles indicate bright spots. Bright spots can be detected as bright spots on the fluorescence image. The bright spot positions of the image 905 obtained by combining the images ( 901 , 902 , 903 , 904 ) corresponding to the four types of A, T, G, and C indicate th...
Embodiment 3
[0085] Figure 15 An example of an image alignment method is shown.
[0086] A reference image 1501 is created based on the design information of the board. For example, the reference image can be created by simulation or the like. Here, the reference image refers to an image of the position of the spot as a reference used to align the position coordinates of the spot on the fluorescent image serving as the bright spot. When the alignment is performed only by the positional information of the spots, the image does not need to be created. The reference image can be prepared in advance according to the substrate used. For the pre-created reference image, the reference image can also be retrieved from the storage medium according to the substrate used. The first reference image created from the design information of the substrate is the position of the adhering speckles of the patterned speckle portion. Depending on the conditions of use, there may be information on the area...
PUM
| Property | Measurement | Unit |
|---|---|---|
| size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com