Genome editing by directed non-homologous DNA insertion using a retroviral integrase-cas9 fusion protein
A technology of retrovirus and fusion protein, applied in the field of genome editing through directional non-homologous DNA insertion using retroviral integrase-Cas9 fusion protein, which can solve the problems of preventing clinical application, off-target mutation, disease, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0325] Example 1: Amplification of Retroviral Integrase-dCas9 Fusion Protein for Editing Mammalian Genomic DNA strong nuclear localization
[0326] Efficient CRISPR-Cas9 editing of mammalian genomic DNA requires nuclear localization of Cas9, a large bacterial RNA-guided endonuclease that normally functions in prokaryotic cells lacking a nuclear membrane. Studies have shown that effective nuclear localization of Cas9 in mammalian cells requires the addition of at least two mammalian nuclear localization signals, one at the N-terminus and one at the C-terminus (Cong et al., 2013, Science 339:819-23) .
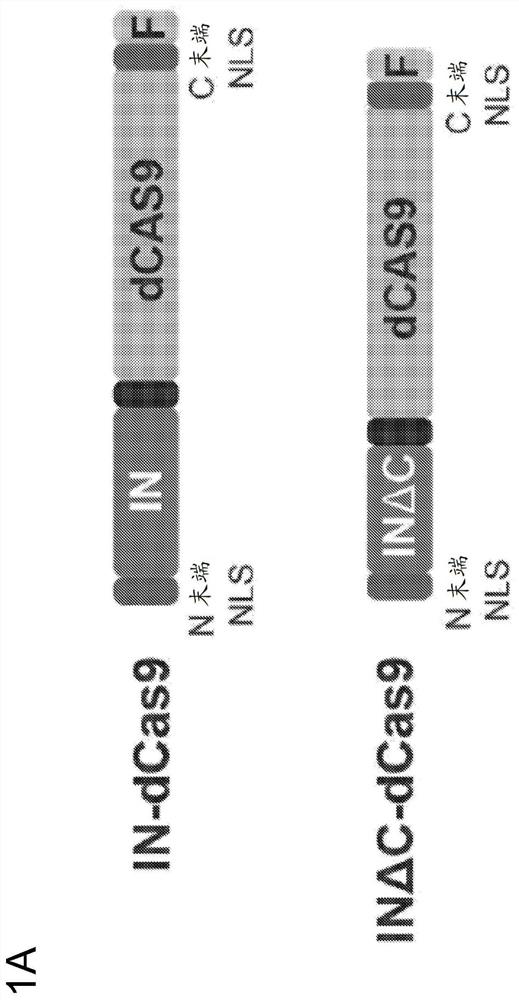
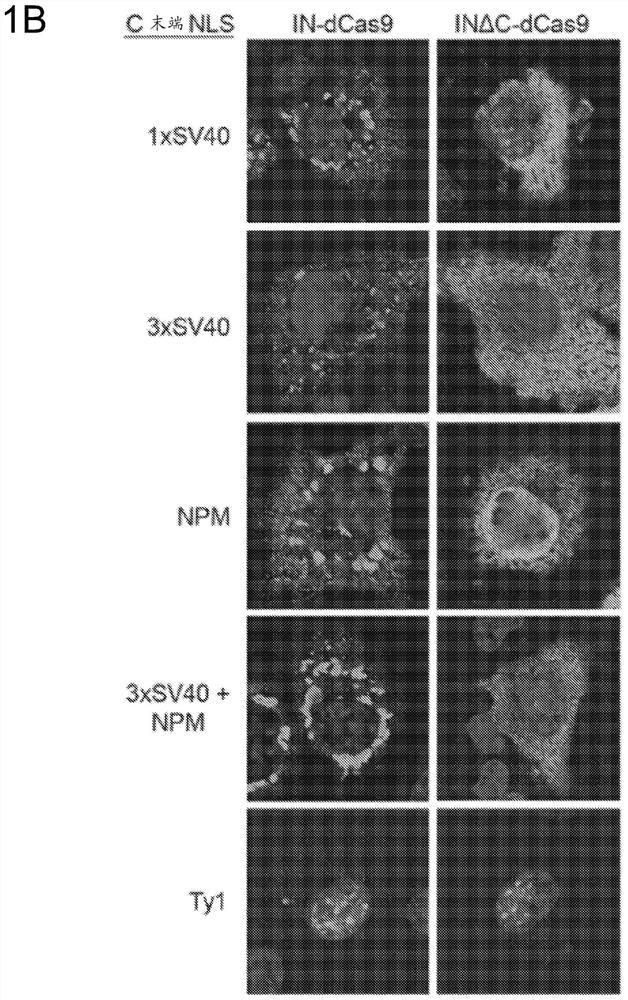
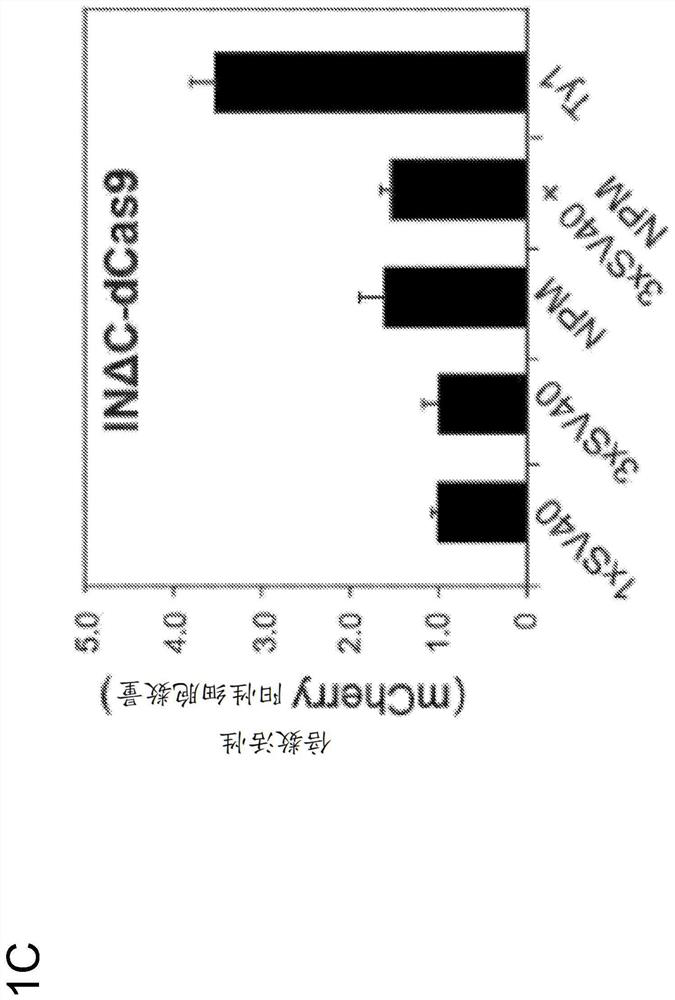
[0327] To facilitate nuclear localization of the retroviral integrase-dCas9 fusion protein for editing, an N-terminal SV40 NLS was included on the integrase in addition to the C-terminal SV40 NLS on dCas9 ( figure 1A). Surprisingly, only a small fraction of the IN-dCas9 fusion protein was nuclear localized when expressed in mammalian cells, as detected using a FLAG antibody...
Embodiment 2
[0331] Example 2: Integrative gene editing approach for correction of muscular dystrophy
[0332] As demonstrated elsewhere herein, the fusion of lentiviral integrases to CRISPR-Cas9 allows sequence-specific integration of large DNA sequences into genomic DNA. This approach can be used to deliver therapeutically beneficial genes to non-pathogenic genomic locations (safe harbor) for permanent correction of human genetic diseases ( figure 2 ). This technique allows sequence-specific integration of large DNA donor sequences containing short viral terminal motifs.
[0333] A major advantage of the gene therapy methods of the present invention is the ability to deliver donor DNA sequences to targeted genomic locations. Furthermore, this approach eliminates the need for homology arms and relies on targeting by guide RNAs, greatly simplifying genome editing. Thus, once a specific reporter donor sequence is generated, it can be directed to any location (or locations) for a varie...
Embodiment 3
[0345] Example 3: Genome Editing - Directed Non-Homologous DNA Integration
[0346] The data presented here demonstrate that optimized integrase-Cas enables efficient editing of mammalian genomes.
[0347] optimized editing
[0348] To optimize IN-mediated integration, determine whether amino acid mutations that enhance integrase catalytic activity, solubility, or interaction with host cell cofactors enhance editing. In addition, the efficiency and fidelity of IN proteins isolated from seven unique classes of retroviruses were assessed.
[0349] To quantify and characterize IN-dCas9-mediated integration in mammalian cells, a plasmid-based reporter system utilizing the blue chromoprotein (amilCP) from Acropora polyporia, when expressed in Escherichia coli, was used. Chromoproteins produce dark blue colonies. Disruption of the amilCP open reading frame abolishes blue protein expression, which can be used as a direct readout for targeting fidelity. In addition, a donor temp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com