Cot DNA suitable for bisulfite sequencing and preparation method and application thereof
A bisulfite and sequencing technology, applied in the field of bisulfite sequencing library, can solve the problems such as the adverse effects of targeted capture, the inability to achieve closed repeat sequences, and the inability to adapt to base changes in bisulfite sequencing libraries. Improve hybridization efficiency and on-target rate, and reduce the effect of impact
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0027] In order to make the object, technical solution and advantages of the present invention clearer, the present invention will be further described in detail below in conjunction with examples. It should be understood that the specific embodiments described here are only used to explain the present invention, not to limit the present invention. The instruments and reagents involved in the examples of this application can be obtained from commercial sources unless otherwise specified.
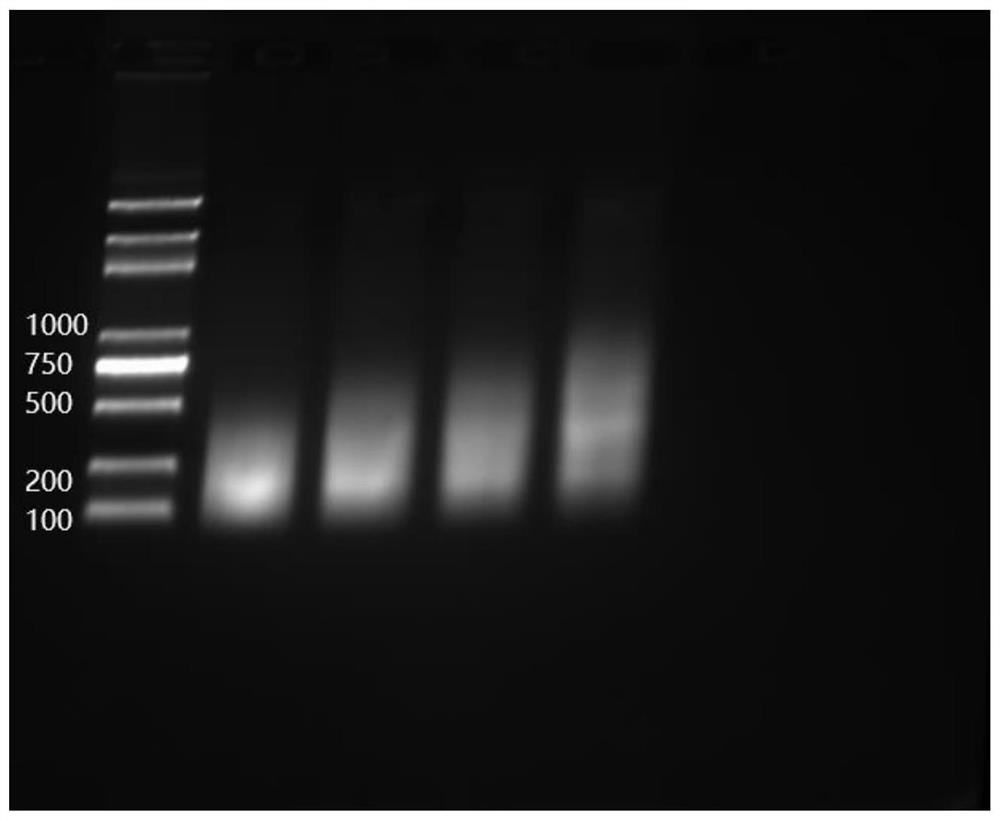
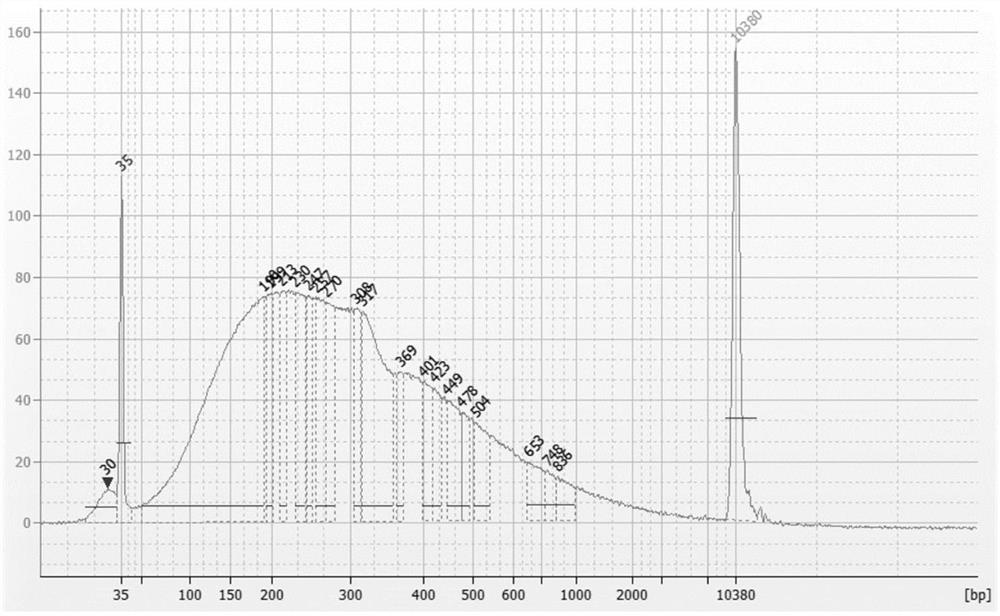
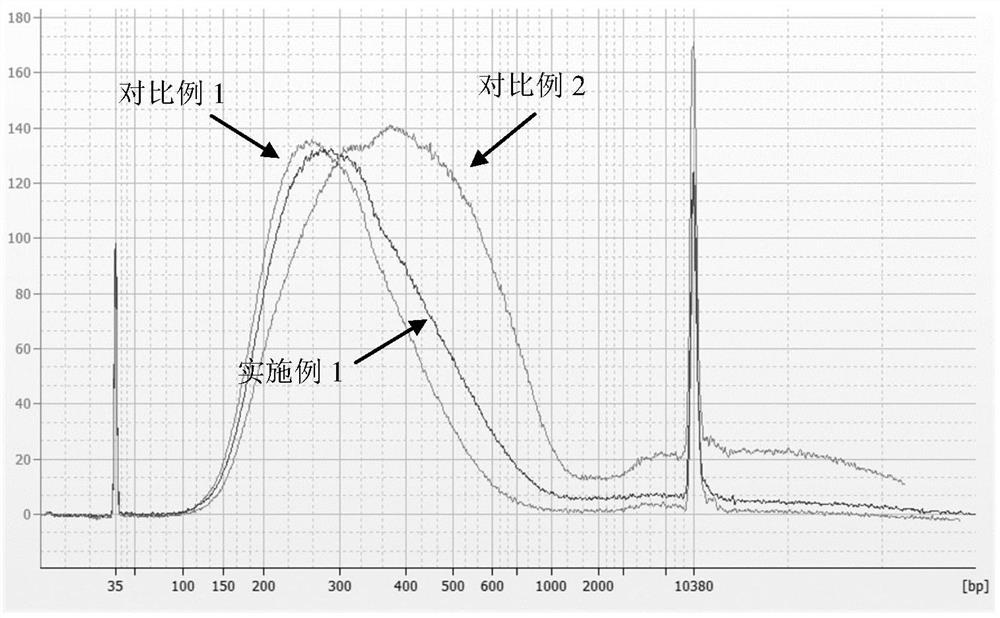
[0028] In this example, taking the human genome as an example, Cot-1 DNA was prepared for bisulfite construction of a sequencing library to block its repeated sequences. The specific process is as follows.
[0029] 1. Sample
[0030] 5 μg DNA samples extracted from human whole blood using QIAamp DNA Blood Kits (Qiagen, 51104).
[0031] 2. Sample fragmentation processing:
[0032] Pre-cool the Covaris S220 non-contact ultrasonic breaker (Chromatin Shearin, USA) to 4-10°C in advance, take 5...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com