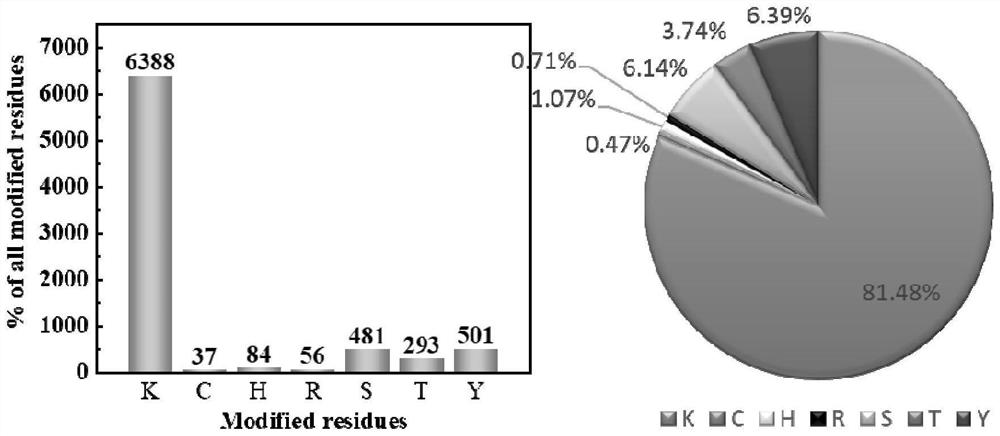
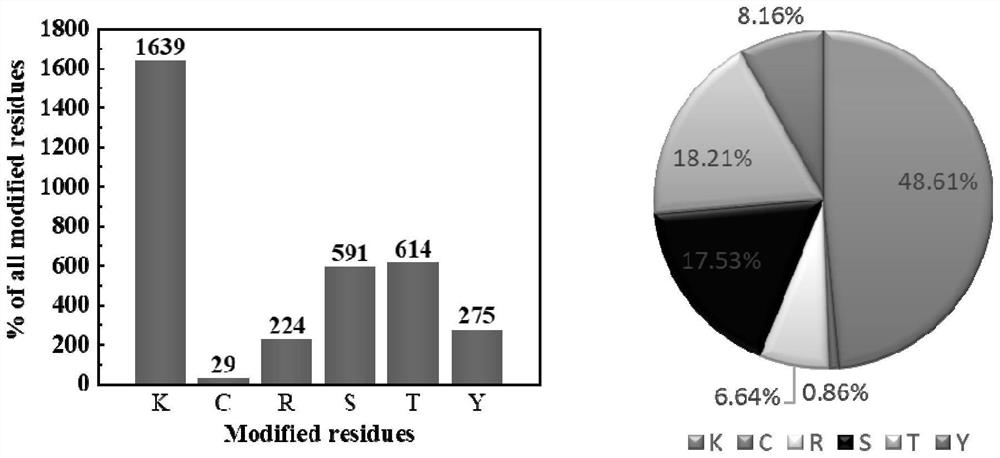
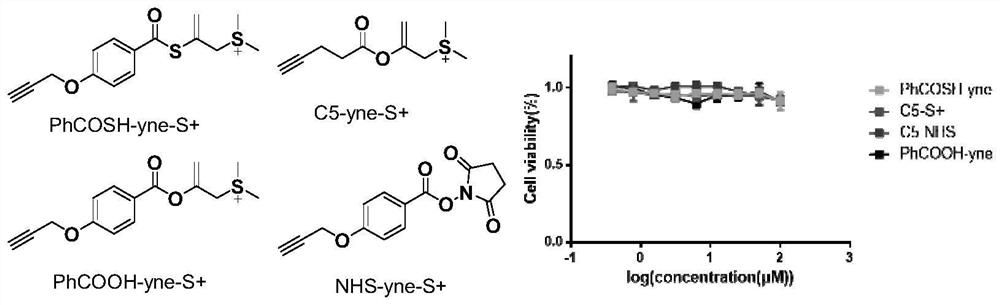
Method for marking protein
A protein and labeling technology, applied in the field of biochemistry, can solve the problems of poor lysine modification selectivity and low solubility of modified substrates, and achieve the effect of significant technological progress.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] 1. Cultivate the A549 cell line in a 10cm cell culture dish until its growth density reaches 80%-90%, discard the medium, add 2mL PBS to wash three times, carefully scrape the cells from the culture dish with a cell scraper, and collect In 2mL ep tube. Use 40% power to sonicate the cells, stop 1s every 1s, and sonicate for 5min.
[0025] 2. Centrifuge at 13000rpm for 10min, collect the supernatant, take 1μL of BSA of 1mg / mL, 2mg / mL, 3mg / mL, 4mg / mL, 5mg / mL and 1μL of the supernatant and place them in a 96-well plate 150 μL of Coomassie Brilliant Blue was added, and the absorbance at 490 nm was measured using a multifunctional microplate reader. Then draw a standard curve of protein concentration and absorbance value, and convert the protein concentration in the supernatant.
[0026] 3. Dilute the protein concentration to 2mg / mL with boric acid buffer solution with pH=10, take 1mL of the lysate and add S-dimethyl-2-propargylsulfonium salt with a final concentration of 5...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com