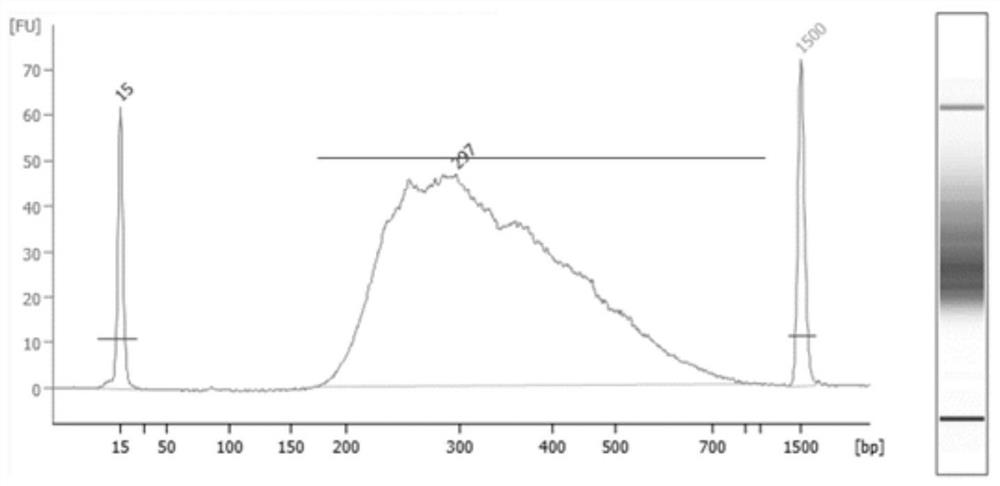
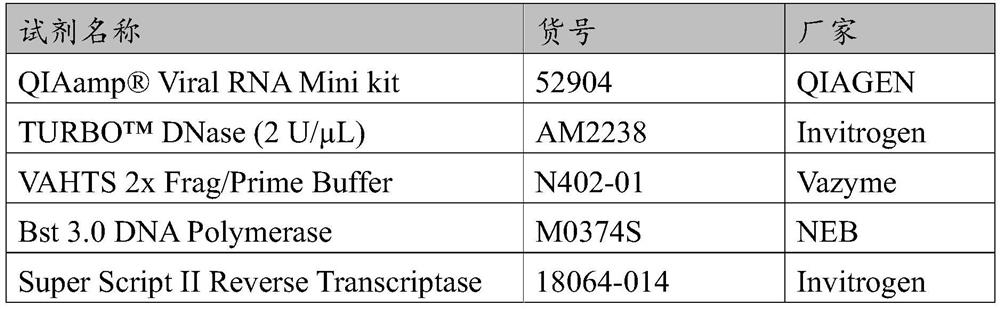
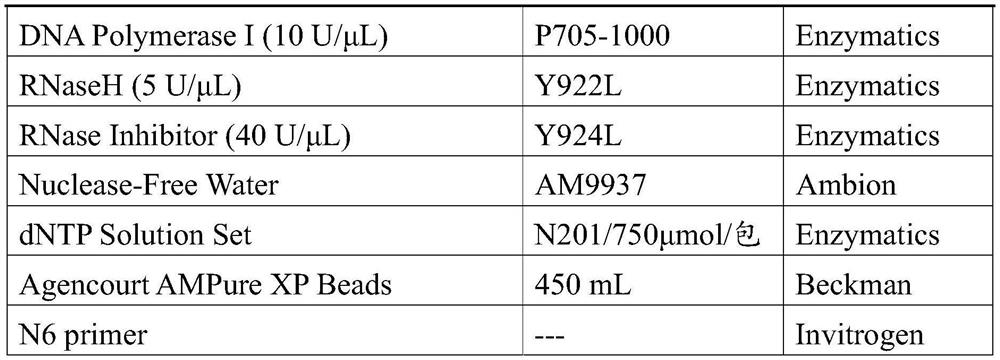
Method for detecting target microbial genome RNA based on high-throughput sequencing technology
A high-throughput, microbial technology, applied in the field of high-throughput sequencing and microbial detection, can solve the problems of low RNA content in microbial genomes, RNA easily degradable background DNA content, and low detection sensitivity, so as to reduce RNA loss and improve microbial genomes. RNA ratio and the effect of improving detection sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0075] Coxsackievirus is an enterovirus whose nucleic acid is single-stranded RNA. This embodiment takes the detection of Coxsackievirus A16 in a sample as an example to illustrate the method for detecting genomic RNA of target microorganisms based on high-throughput sequencing technology of the present invention. The Coxsackievirus A16 type was derived from the remaining clinical samples provided by the applicant's cooperative hospital. Exemplary detection steps of the method of the present invention are described below.
[0076] 1. Coxsackievirus A16 mock sample preparation: add Hela cells to normal saline to prepare 10 5 cell / mL solution of Hela cells, the resulting solution was divided into four equal parts, and Coxsackie virus A16 type was added to the viral load of 1000cp / mL, 500cp / mL, 100cp / mL, 50cp / mL respectively, and the obtained solution containing different Coxsackievirus type A16 mock sample of viral load.
[0077] 2. Nucleic acid extraction: use The Viral RN...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com