Method for detecting nucleic acid marker RNA virus based on CRISPR/Cas12a system innovative activation mode
A technology of RNA virus and activation method, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve problems such as low RNA content, complex operation process, and cumbersome operation process.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0035] The present invention will be further described in the following examples, but the present invention is not limited thereto.
[0036] Concrete steps of the present invention are as follows:
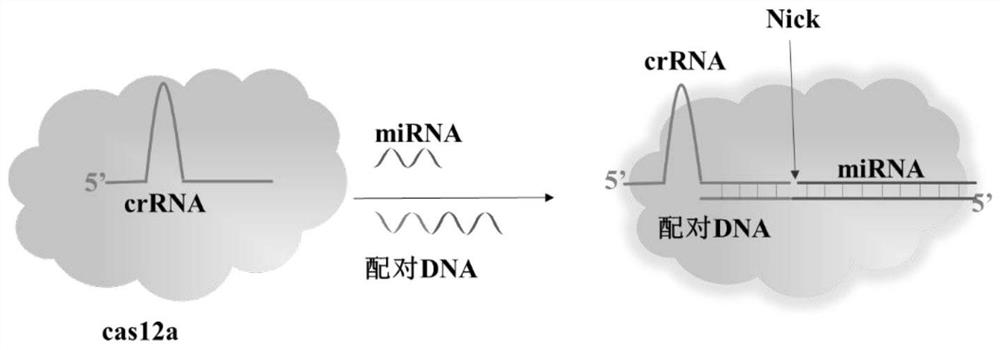
[0037] (1) Design crRNA and paired DNA
[0038] Design primers on the official website of Integrated DNA Technologies (IDT) in the United States:
[0039] Cr RNA: 5'UAAUUUCUACUAAGUGUAGAUCGUCGCCGUC3'
[0040] Paired DNA: 5'TCAACATCAGTCTGATAAGCTAGACGGCGACG3'
[0041] miRNA-21: 5'UAGCUUAUCAGACUGAUGUUGA3'.
[0042] (2) Construction of CRISPR / Cas12a system
[0043] Add cas12a enzyme and 1x NEB 2.1 reaction buffer, double distilled water, 1nM paired DNA, molecular beacon FQ and designed crRNA (or designed paired DNA) into a 100 μl EP tube.
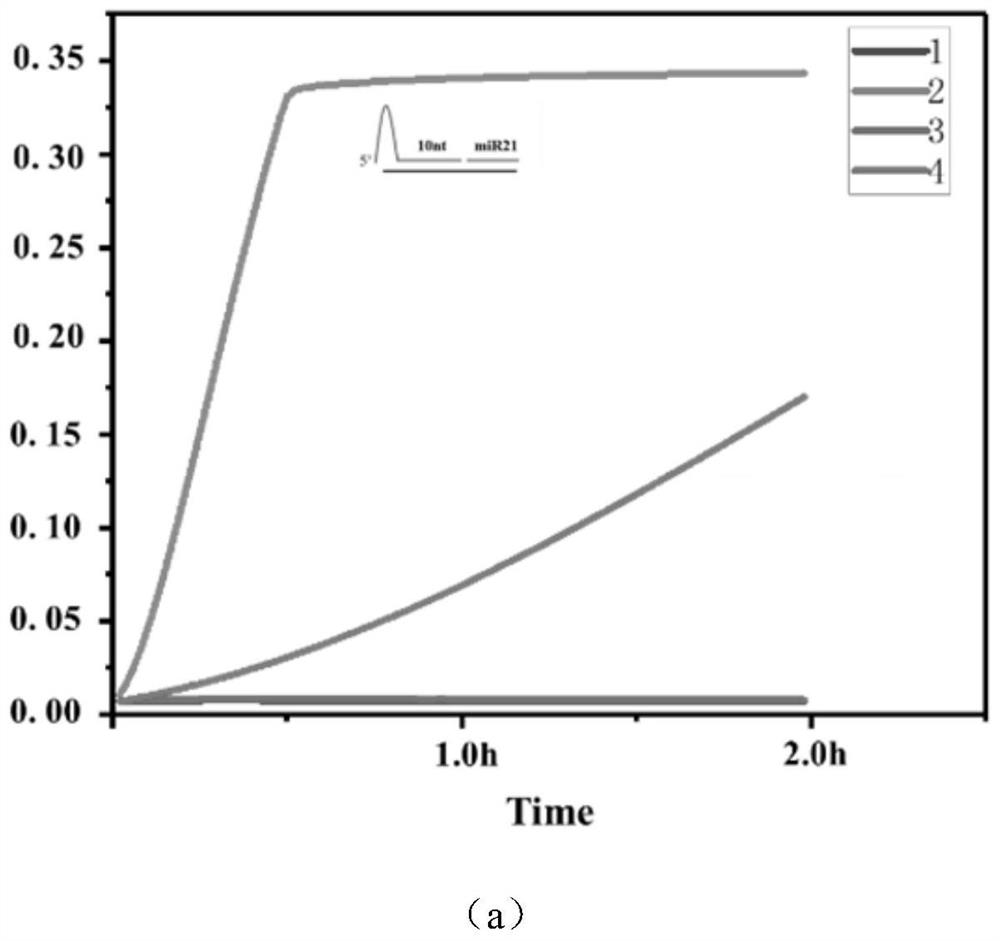
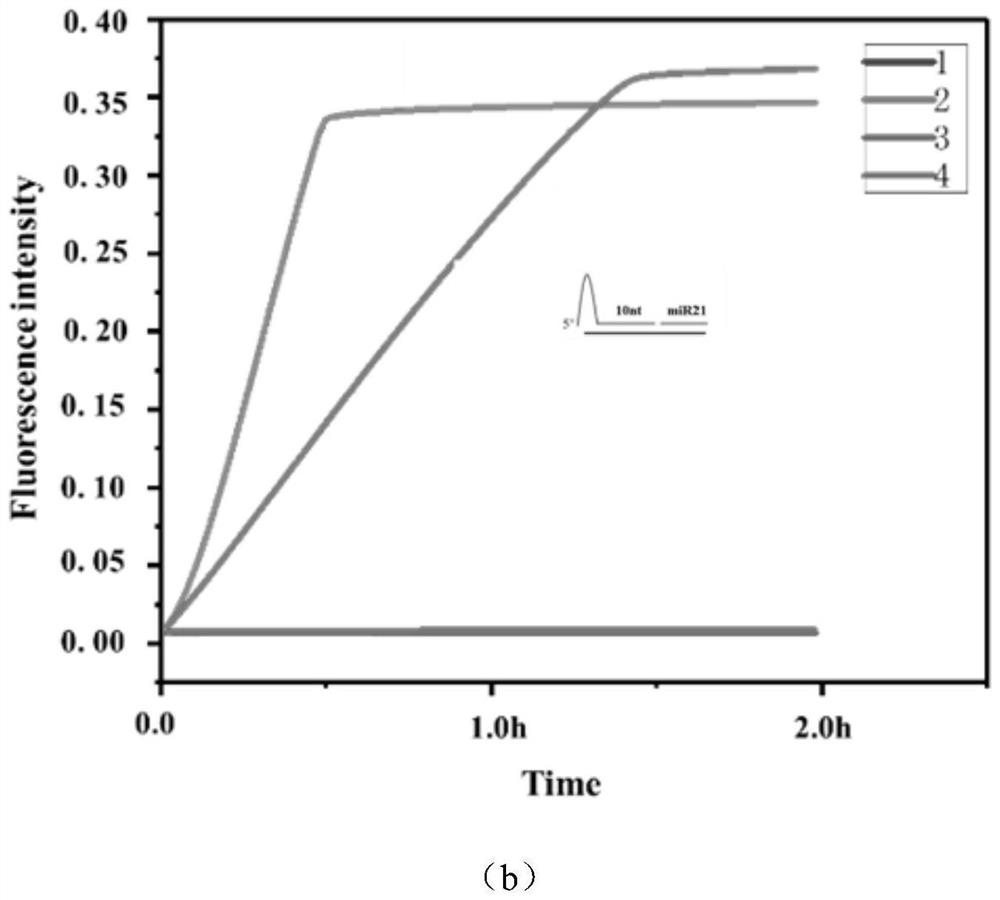
[0044] (3) Add 1 μM target miRNA-21
[0045] Add target RNA to detect fluorescence changes in real-time fluorescent quantitative PCR.
[0046] (4) Experiment optimization and data processing
[0047] The design of crRNA, the design of paired ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com