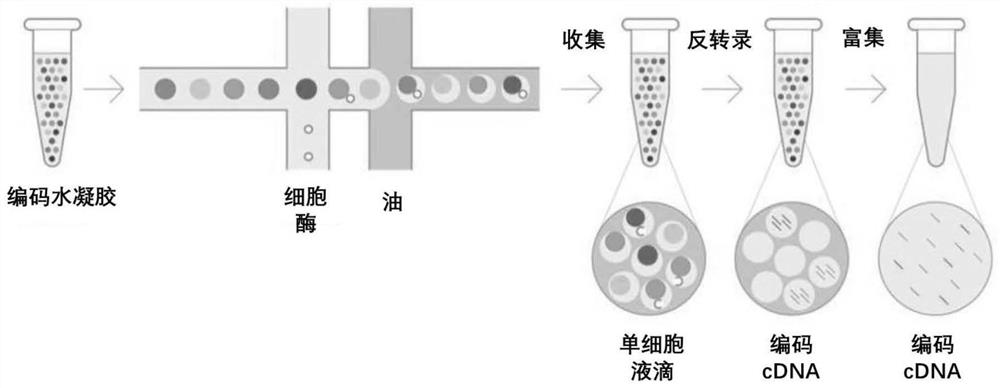
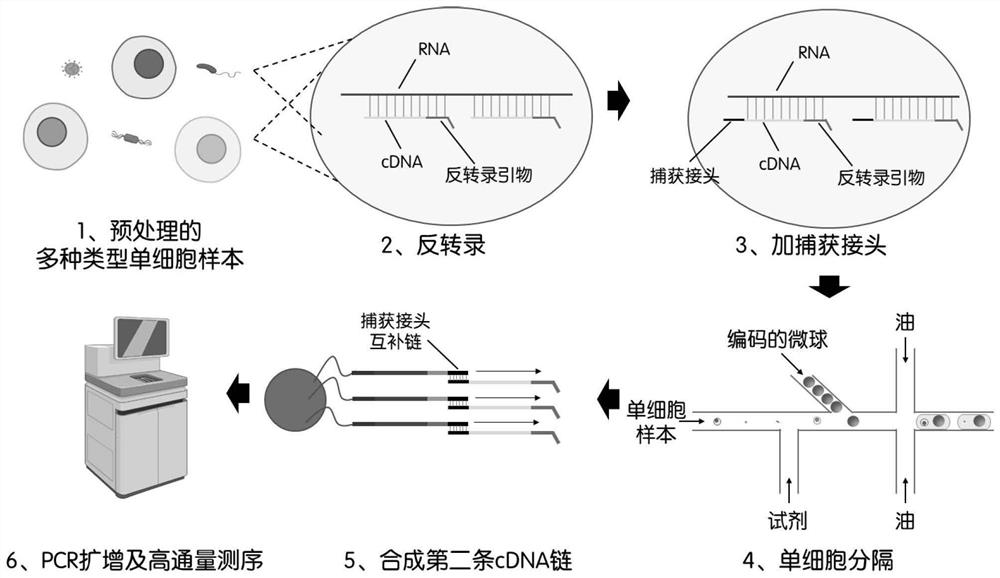
Single cell transcriptome sequencing method and application thereof
A transcriptome sequencing and single-cell technology, applied in biochemical equipment and methods, combinatorial chemistry, microbial measurement/inspection, etc., can solve the problems of low sensitivity, undetectable, poor sequencing effect, etc., and achieve RNA detection sensitivity high effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach
[0056] In order to make the purposes, technical solutions and advantages of the embodiments of the present invention clearer, the technical solutions in the embodiments of the present invention will be clearly and completely described below with reference to the accompanying drawings in the embodiments of the present invention. Obviously, the described embodiments These are some embodiments of the present invention, but not all embodiments.
[0057] Based on the embodiments of the present invention, all other embodiments obtained by those of ordinary skill in the art without creative work fall within the protection scope of the present invention.
[0058] The technical solutions of the present invention are further described below with reference to the accompanying drawings and through specific embodiments.
Embodiment 1
[0059] Example 1: Sequencing of E. coli samples
[0060] sample preprocessing
[0061] Take about 1 million different strains (1, 2, 3, 4) of Escherichia coli to verify that the patent of the present invention can be used for single-cell transcriptome sequencing of bacterial samples. ) PBS buffer (PBST) was washed three times, and the agglomerated bacteria were removed by shaking and filtration, so that the bacteria in the sample formed a single bacterial suspension, and 1% paraformaldehyde (purchased from Beijing Soleibao Technology Co., Ltd.) was added. Fix overnight at 4°C. The fixed bacterial samples were washed three times with PBST buffer, lysozyme (purchased from Thermo Fisher Scientific, USA) was added to lyse the cell wall, treated at 37°C for 15 minutes, and washed three times by adding PBST buffer.
[0062] Reverse Transcription
[0063] The pretreated samples were added with reverse transcriptase (purchased from Thermo Fisher Scientific, USA), reverse tran...
Embodiment 2
[0073] Example 2: Sequencing of mixed samples of Escherichia coli and Bacillus subtilis
[0074] About 1 million mixed samples of Escherichia coli and Bacillus subtilis were taken to verify the accuracy of single-cell transcriptome sequencing using the patent of the present invention. Sample pretreatment, reverse transcription, addition of capture adapters, single cell isolation, synthesis of the second strand of cDNA, library construction, and high-throughput sequencing were performed using the methods described in Example 1 above. The sequencing results showed that a total of 251 bacteria were detected in the mixed samples of Escherichia coli and Bacillus subtilis after genome alignment, and 4 of them contained both Escherichia coli cDNA and Bacillus subtilis cDNA. The remaining bacteria contain almost only Escherichia coli cDNA or Bacillus subtilis cDNA, and the overall contamination rate is probably less than 2% (see Figure 5 ). It shows that the application of the pa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com