Method for detecting transgenic event in plant
A technology of genetically modified events and events, applied in the fields of biotechnology and agricultural science, can solve the problems of cumbersome and complicated operation steps, high reagent and labor costs, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1 Detection of the number of transgenic events
[0037] Plant softening treatment
[0038] 1. Transfer the sample to a petri dish and dry it in a constant temperature oven at 37°C for 48h
[0039] 2. The sample is placed in the tissue grinder, and the tissue powder is collected after sufficient grinding
[0040] 3. Take 1g of powder sample, mix well, transfer it into EP tube and wrap the tissue powder with 3% agar, and put it into the agar after it cools and solidifies.
[0041] Fix in FAA fixative solution (50% concentration for young plants and 70% concentration for old hard tissues) for 24h.
[0042] 4. Take out running water and rinse for 2-4h
[0043] 5. For long-term storage, it can be transferred back to FAA fixative
[0044] Paraffin-embedded sections of samples 1. Materials: fresh tissues were fixed in FAA fixative for more than 24 hours. Take the tissue out of the fixative solution, trim the tissue at the target site with a scalpel in a fume hoo...
Embodiment 2
[0062] Example 2 Detection of Sensitivity
[0063] According to the experimental method in Example 1, prepare rapeseed A seeds containing only PAT / bar transgenic events, B rape seeds containing only CP4-EPSPS transgenic events, and C rapeseed seeds containing both CP4-EPSPS and PAT / bar transgenic events seed. The three kinds of rapeseed seeds ABC were mixed with non-transgenic rapeseed seeds at a ratio of 0.1%, and then tested.
[0064] The results showed that in the case of only 0.1% of transgenic seeds, the fluorescent immunoassay method could accurately identify the number of transgenes in the sample to be tested, and its detection sensitivity was 0.1%.
Embodiment 3
[0065] Example 3 Sensitivity detection after shielding autofluorescence treatment
[0066] The operation is performed according to the experimental method in Example 1-2. Specifically, after antigen retrieval, the operation of shielding autofluorescence is performed. The specific operation is as follows: adding autofluorescence quencher A and autofluorescence quencher B in sequence, and finally performing The serum is blocked; the autofluorescence quencher A is a mixture of pH 7.4 phosphate buffer and sodium borohydride; the autofluorescence quencher B is a mixture of Sudan black B and absolute ethanol; autofluorescence is added first Quencher A, incubate at room temperature for 10-20 min, and wash; then, add autofluorescence quencher B, incubate at room temperature for 10-15 min, wash, and then perform immunofluorescence staining and observation.
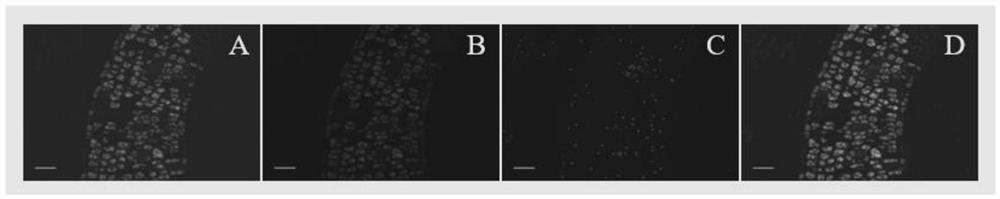
[0067] The three rapeseed seeds ABC were mixed at a lower ratio in non-transgenic seeds and subjected to a shielded autofluores...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com