Method for predicting splicing sites with paired two ends
A splice site and prediction method technology, applied in the field of double-ended paired splice site prediction, to facilitate the research of double-ended paired splice sites, increase convenience, and improve prediction performance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
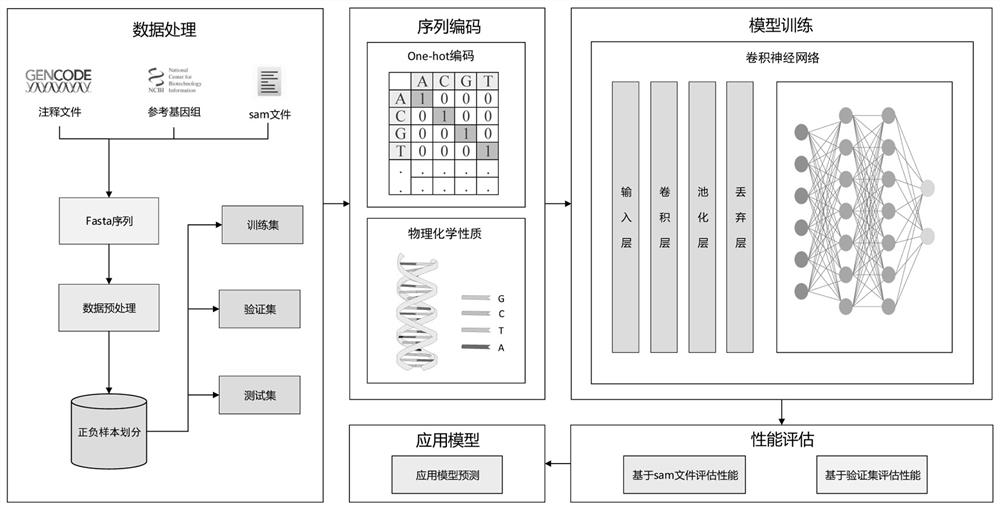
[0055] Such as figure 1 As shown, a double-ended paired splice site prediction method includes the following steps:
[0056] 1) Use the human reference genome sequence as the source, and collect the splice site sequence data according to the reference genome sequence file and the reference genome annotation file. Specifically, to collect the human splice site data set, you first need to download the human reference genome sequence from the NCBI database, and then download it from The GenCode database downloads the reference genome annotation file, and combines the reference genome sequence and annotation file to obtain the required information.
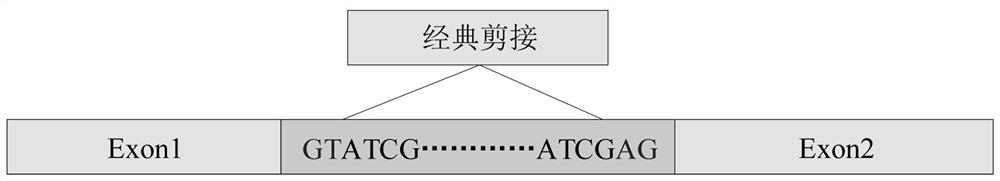
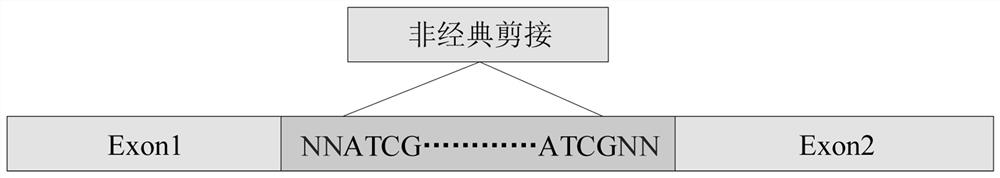
[0057] The splice site sequence data includes canonical splice site sequences and non-canonical splice site sequences such as figure 2 and image 3 As shown, data processing is performed on the collected splice site sequence data, including the length of the data, introns and exons for region identification processing, and after po...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com