Method for determining TAT cells
A TAT-TCR, cell technology, applied in the field of determining TAT cells in blood, can solve the problems of cumbersome experiments, low throughput, and no way to cover T cells.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0087] Example 1: Determination of TAT cells and TAT-TCR sequences
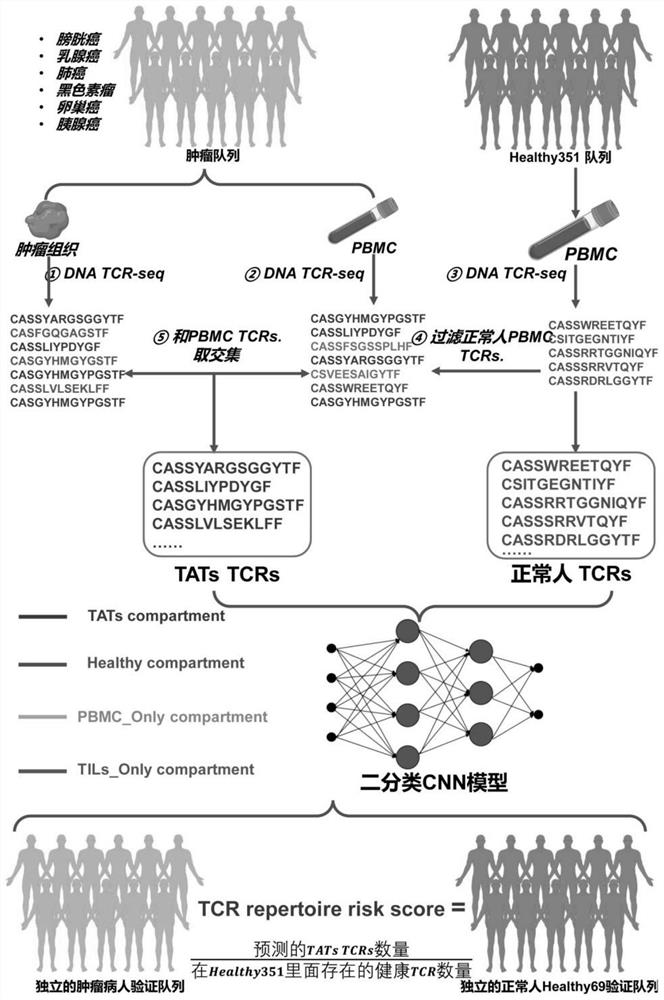
[0088] The inventors collected the TCR sequencing data of more than 100 cases of tumor tissue / peripheral blood pairings from the ImmuneACCESS public database, and divided all TCR clones into different subgroups based on the CDR3 sequence information. Subsequently, the inventors collected TCR sequencing data of more than 500 PBMC samples from normal healthy people from the ImmuneACCESS database. The TCR sequences present in the above-mentioned normal healthy human PBMC samples were excluded from the TCR sequences present in both PBMC (peripheral blood mononuclear cells) and TIL (tumor infiltrating T cells), and the resulting TCR sequences were defined as TAT-TCR sequences . T cells containing the TAT-TCR sequence were defined as TAT cells ( figure 1 ).
Embodiment 2
[0089] Example 2: Assessment of Cancer Risk Using TAT-TCR Sequences
[0090] The inventors followed the data filtering and screening steps as shown in the figure below ( figure 1 ), a TAT-TCR-based binary prediction model was constructed to predict the probability that each TCR is a tumor-related TCR.
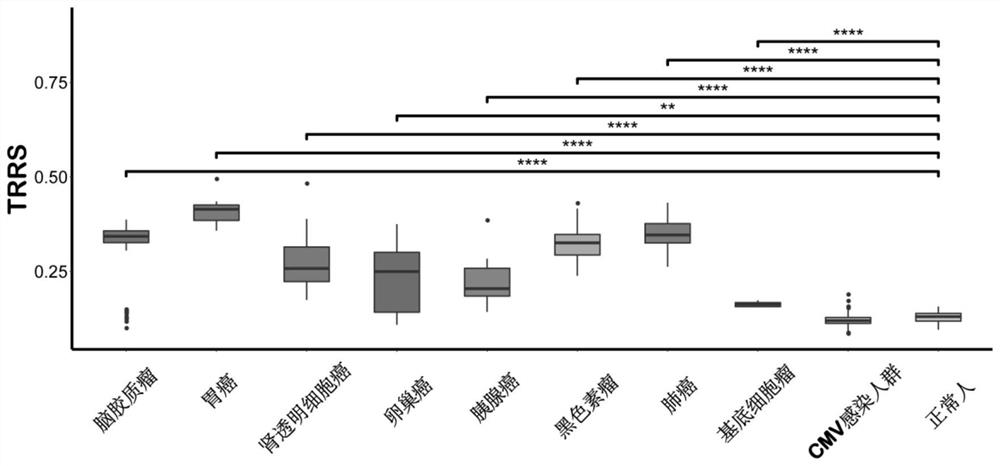
[0091] Subsequently, the inventor designed a TCR Repertoire Risk Score (TRRS) to quantitatively evaluate the enrichment degree of tumor-related T lymphocytes in the peripheral blood of patients. The specific calculation method is as follows: for each patient sample, the inventor uses the constructed The binary classification model predicts each TCR sequence detected in its PBMC, and obtains the probability that the TCR sequence is TAT; then under a given probability threshold (66% or 75%), we calculate that the patient is predicted as The number of TAT sequences a; next, we calculate the number b of TCR sequences (obtained from the above-mentioned public database) in the PBM...
Embodiment 3
[0093] Example 3: Analysis of clonal proliferation ability of TAT cells
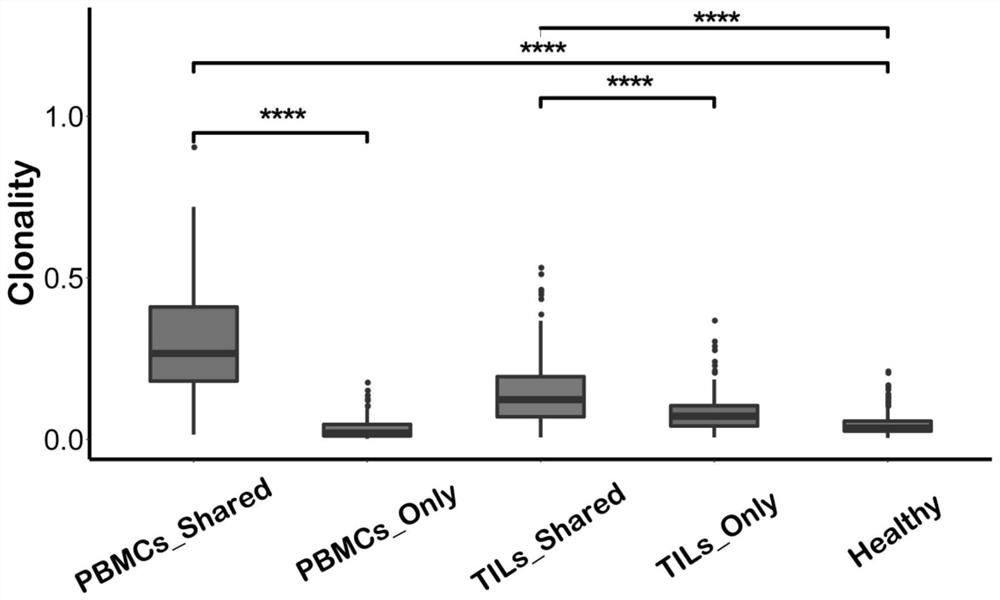
[0094] The inventors analyzed the clonal proliferation of different T cell subsets by using the LymphoSeq R package (https: / / github.com / davidcoffey / LymphoSeq).
[0095] The inventors found that the clonal proliferation of T-cell subsets that share TCR sequences co-existing in the microenvironment of blood and tumor infiltration is higher, especially the clonal proliferation of such TAT cell subsets derived from PBMC is higher, indicating that the clonal proliferation is higher. This type of T cell subsets (TAT cells) have stronger binding ability to tumor-associated antigens and have stronger tumor-killing activity.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com