SNP (Single Nucleotide Polymorphism) molecular marker for sex identification of silver arowana and application thereof
A molecular marker, silver dragon fish technology, applied in the direction of recombinant DNA technology, microbial measurement/inspection, DNA/RNA fragments, etc., can solve problems such as limiting reproductive efficiency and success rate, inability to penetrate scales, and gender indistinguishability, etc. Achieve important economic value and social value, solve major industrial problems, and improve reproductive efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] In this example, a high-throughput sequencing strategy was used to select 5 female dragon fish samples and 5 male fish samples for DNA extraction. According to the requirements of the Illumina library construction process, a double-ended genomic DNA library with an insert size of 500 bp was constructed. Then high-throughput sequencing was performed on the genome using the Illumina NovaSeq sequencing platform, with a sequencing volume of 30 Gb per sample and a sequencing strategy of Pair-End 150 bp.
[0025] Using a high-throughput sequencing sequence alignment strategy, the single nucleotide polymorphism sites (SNPs) of the sequenced female samples and the single nucleotide polymorphism sites (SNPs) of the male samples were detected, and the female samples were screened. are identical between male and female samples, and the different SNP loci of male and female samples are used as molecular markers for sex identification.
[0026] The female samples and male samples we...
Embodiment 2
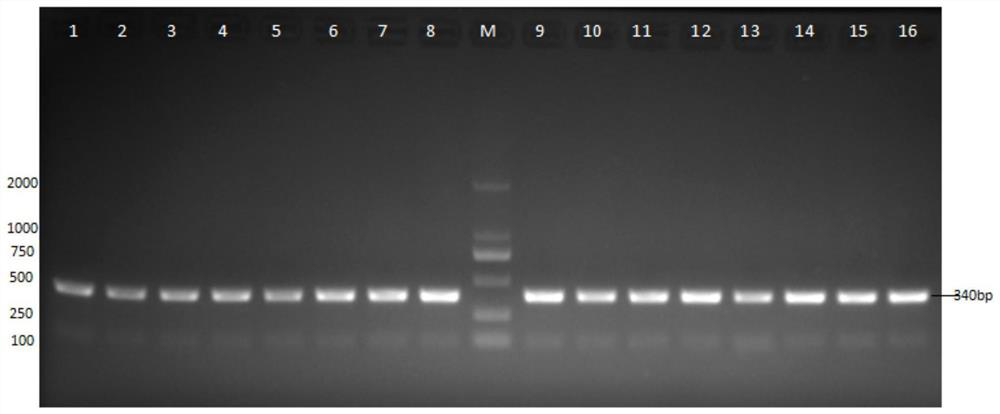
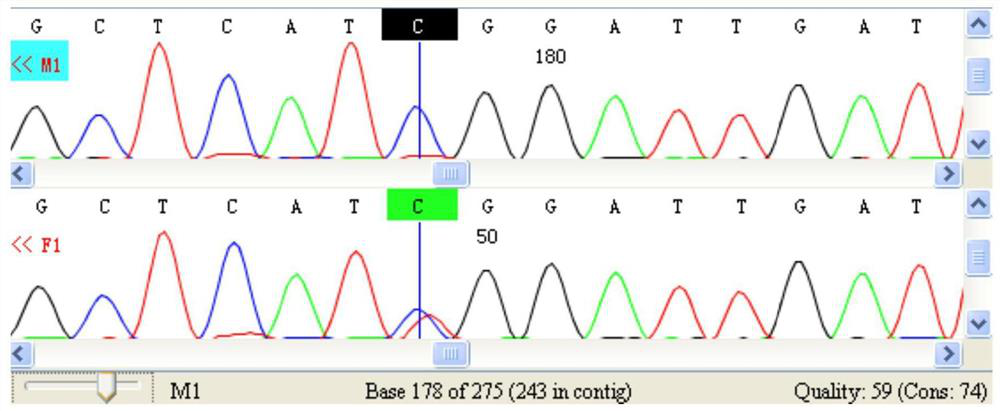
[0035] In order to verify the accuracy of the SNP site to identify the gender of Arowana, this example carried out the verification of the SNP molecular markers in Example 1, and verified whether the molecular markers detected by Arowana were accurate by PCR and Sanger sequencing. Specifically, genomic DNA was extracted from the non-high-throughput sequencing samples, the specificity of the primers was verified by PCR and Sanger sequencing, and the physiological sex of the silver arowana samples was compared with each other to confirm the SNP molecular markers in Example 1 , Whether the primers can be used for gender identification of silver arowana.
[0036] 1. Sample preparation
[0037] Select 60 silver arowana samples, including 30 female samples and 30 male samples, for verification. The following table shows the sample information:
[0038] Experimental verification samples
[0039]
[0040]
[0041]
[0042] 2. Genomic DNA extraction
[0043] Extraction was ca...
Embodiment 3
[0062] The present embodiment provides a method for gender identification of silver arowana, comprising the following steps:
[0063] (1) extracting the genomic DNA of the silver arowana to be tested;
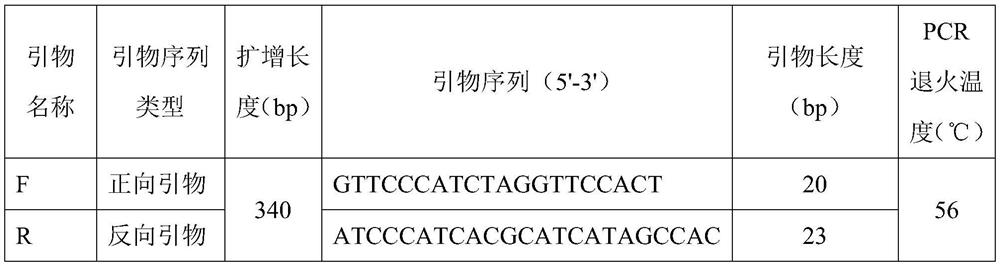
[0064] (2) using the genomic DNA of the silver arowana to be tested as a template, using the upstream primer F and the downstream primer R whose sequences are shown in SEQ ID NO.2 and SEQ ID NO.3, respectively, to carry out PCR amplification reaction;
[0065] (3) After the reaction is completed, the Sanger sequencing is combined for analysis to determine the genotype of the sample, and the gender is identified according to the 243rd position in the sequence SEQ ID NO.1.
[0066] Specifically, the reaction system and amplification procedure used in the PCR amplification reaction in step (2) are the same as those in Example 2.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com