HIL7/hCCL19 double-gene recombinant oncolytic virus as well as preparation method and application thereof
An oncolytic virus and genome technology, applied in the field of genetic engineering, can solve the problems of reduced tumor sensitivity, lower virus killing and replication ability than wild-type virus, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0092] Example 1 Construction of recombinant virus
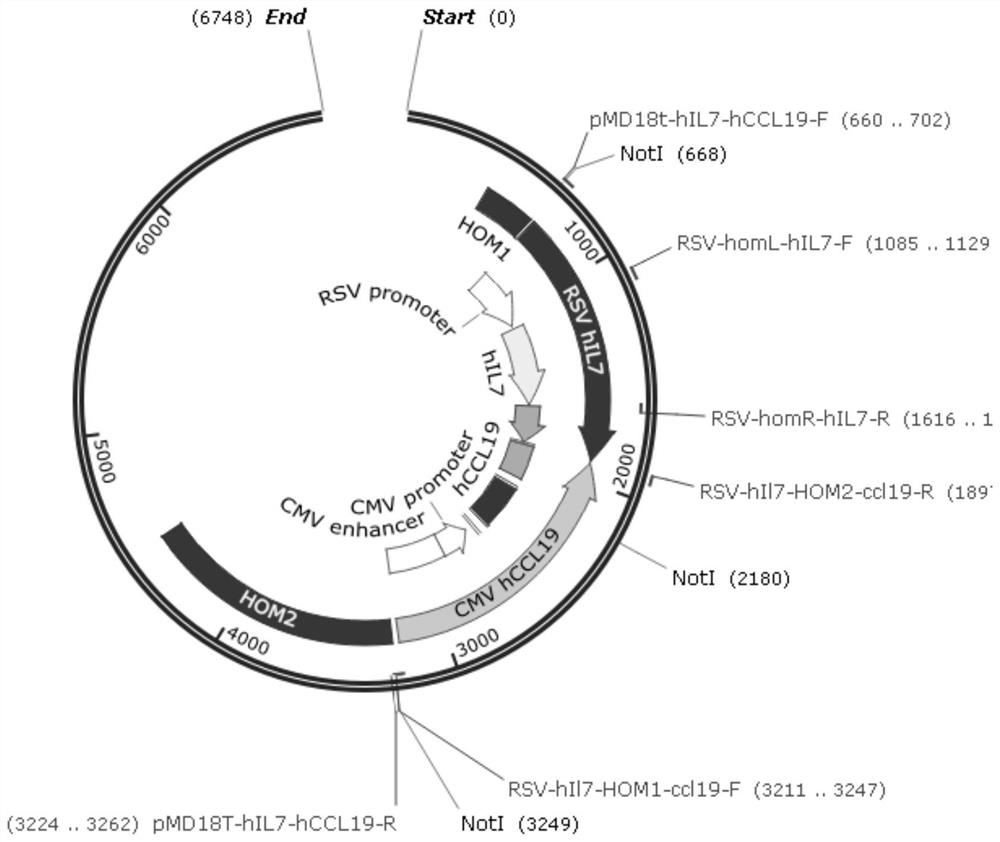
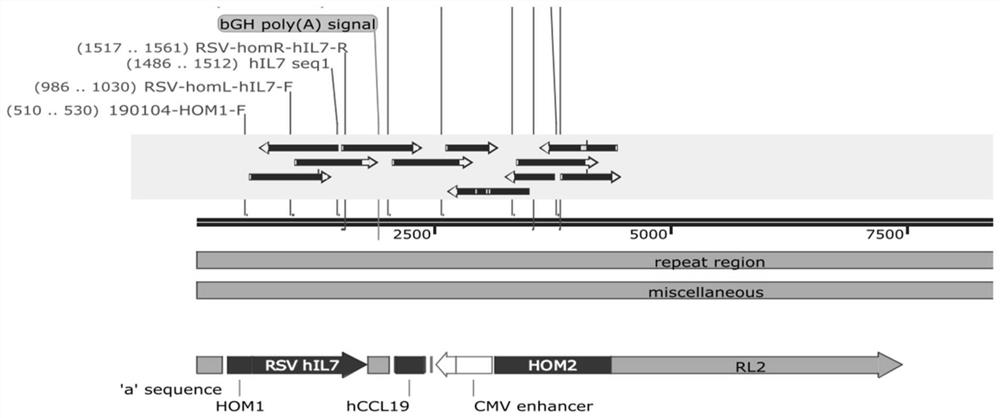
[0093] Insert the gene sequences encoding hIL7 and hCCL19 between the homology arm sequences on the left and right sides of the ICP34.5 gene to obtain the pMD18T-HOM-hIL7-hCCL19 donor plasmid, as shown in the appendix. figure 1 , the ICP34.5 gene of the HSV-1 / ICP47-knockout virus strain was replaced by the therapeutic genes hIL7 (human IL7) and hCCL19 (human CCL19) by the Crisper-Cas9 system, the recombinant virus was constructed, and the successfully constructed virus was screened. The first nucleotide of the start codon of the ICP34.5 gene is used as the first nucleotide for sequence coding, and the gene insertion position is between the 134th nucleotide of the start codon and the 160th nucleotide downstream of the stop codon. between nt522-nt1271 and nt124324-nt125073 in the wild-type HSV-1 genome (GeneBank: JQ780693.1).
[0094] Using the Crisper-Cas9 gene editing system to cut the Target DNA (targeted DNA) of the oncol...
Embodiment 2
[0107] Example 2 PCR verification
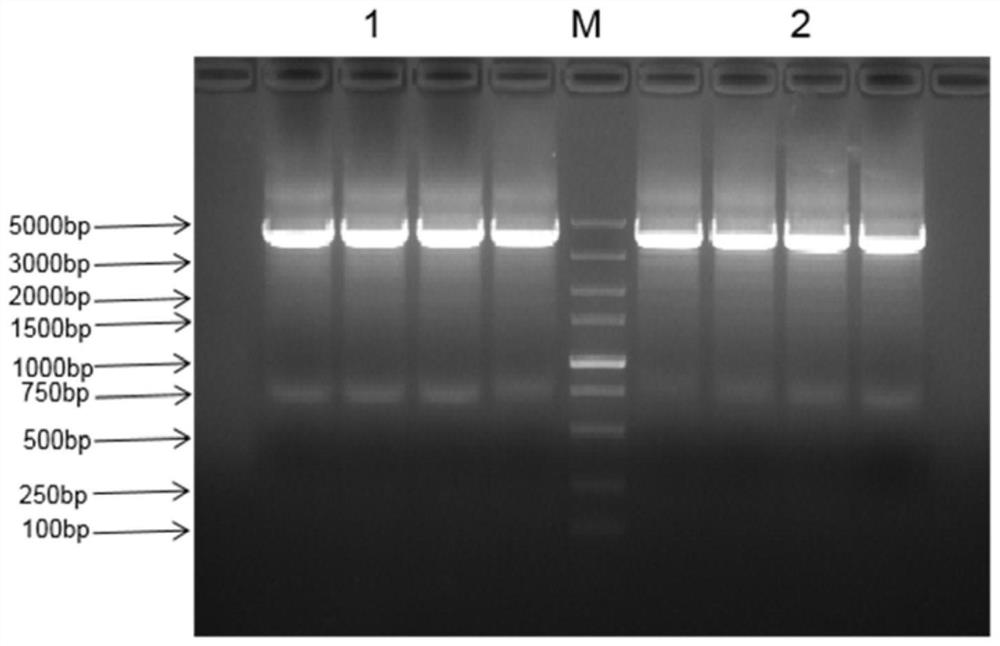
[0108] According to the virus titer, the virus (recombinant virus obtained in Example 1) was infected with VERO cells covered with six-well plates, the size of virus plaques was observed, and DMEM medium containing 0.01% neutral red was added for staining. Monoclonal virus plaques were amplified and the genome was extracted for PCR verification.
[0109] (1) Experimental materials:
[0110] Virus monoclonal Δ47-hIL7-hCCL19-1 and Δ47-hIL7-hCCL19-2 after multiple rounds of screening.
[0111] (2) PCR verification: the primer is HOM1-F / 34.5-flank-seq, the PCR product size is 3975bp,
[0112] HOM1-F: 5'-CGCACAGTCCCAGGTAACCTC-3', as shown in SEQ ID NO:8;
[0113] 34. 5-flank-seq: 5'-GGCGTGTCTCTGTGTATGAGTCAGGGGGTCC-3' as shown in SEQ ID NO:9.
[0114] PCR system
[0115]
[0116]
[0117] PCR program: 98°C, 3min; (98°C, 10s; 55°C, 15s; 68°C, 3min) x 35 cycles; 72°C, 5min; 12°C, forever.
[0118] (3) Sequencing results and ana...
Embodiment 3
[0119] Example 3 Protein ELISA identification
[0120] (1) Experimental materials:
[0121] The experimental group Δ47-hIL7-hCCL19 refers to the Δ47-hIL7-hCCL19-1 recombinant oncolytic virus, and the negative control is the Δ47 oncolytic virus.
[0122] (2) ELISA verification: Vero (African green monkey kidney cells) was used as the host cell, and the conditions were as follows: Δ47-hIL7-hCCL19, Δ47 infection MOI (ratio of virus to cell number) was 0.01, after infection with Vero cells, 24 , 48, 72 hours to collect the cultured virus culture supernatant.
[0123] (3) hCCL19 expression results:
[0124] The expression results of hCCL19 in Vero cells are shown in Table 1.
[0125] Table 1:
[0126]
[0127] The expression results of hCCL19 in Vero cells are as follows Figure 4 As shown, the expression of hCCL19 protein at three time points (24h, 48h, 72h) is shown. The figure shows that with the increase of infection time, the expression of hCCL19 first increases and th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com