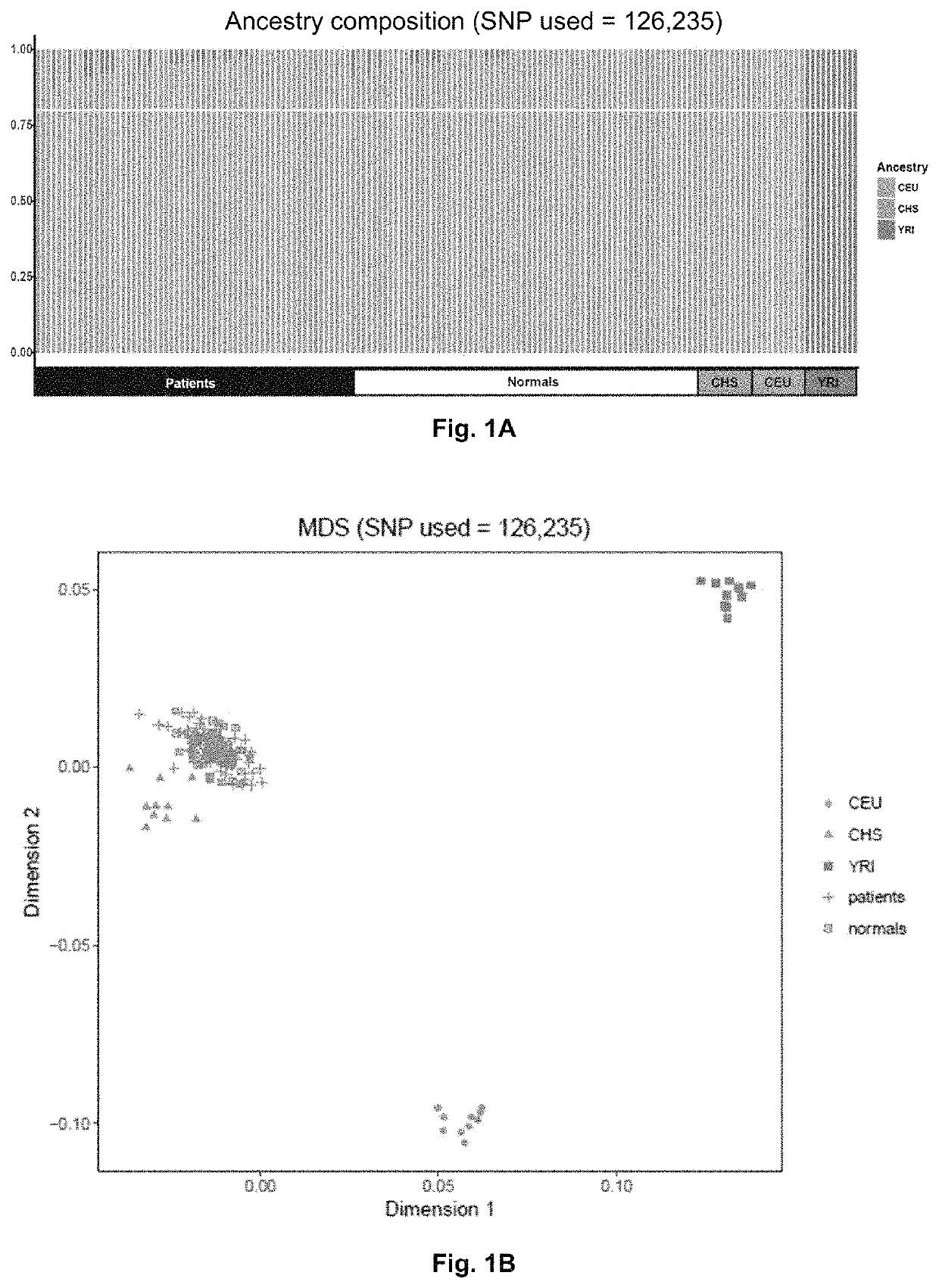
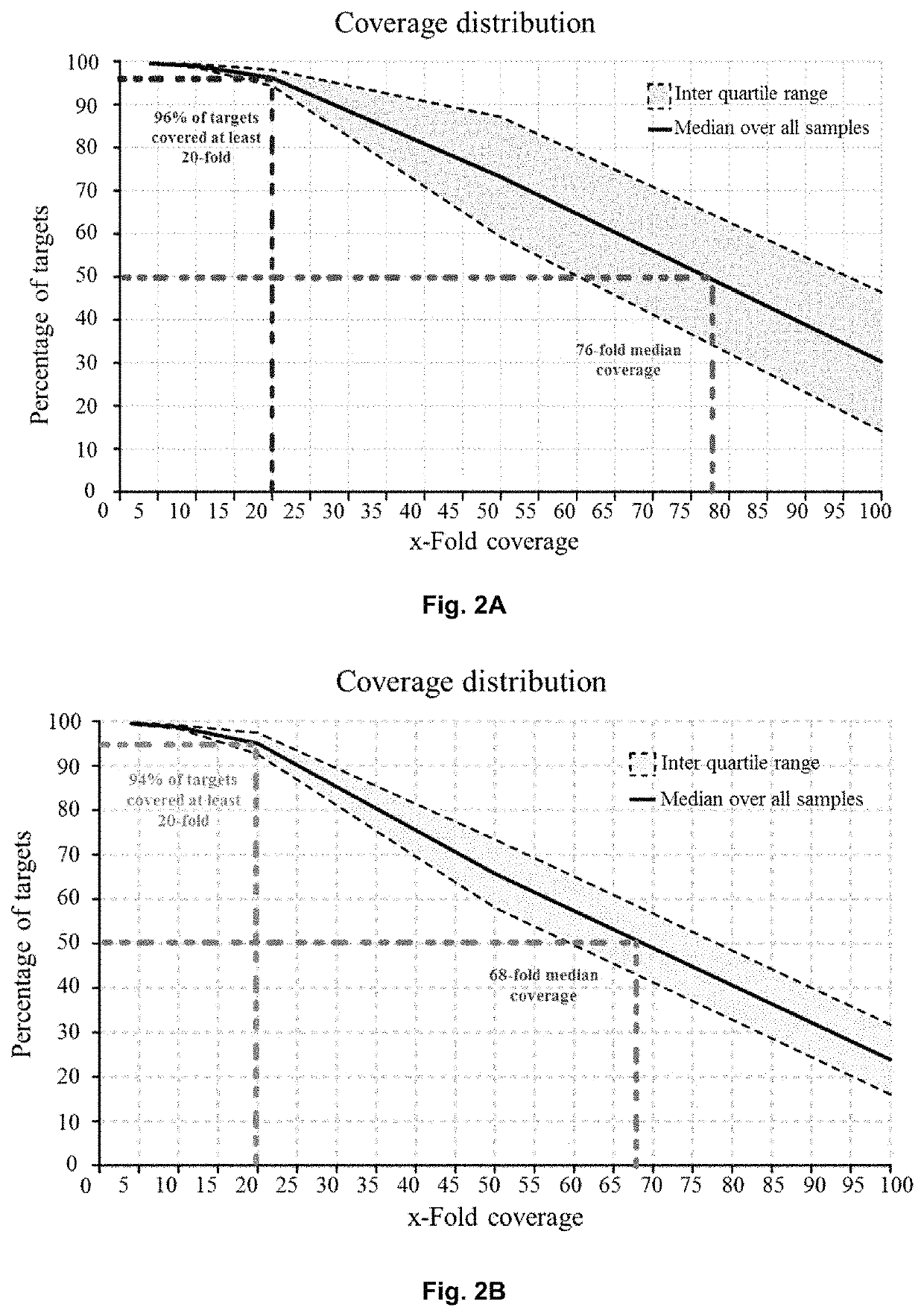
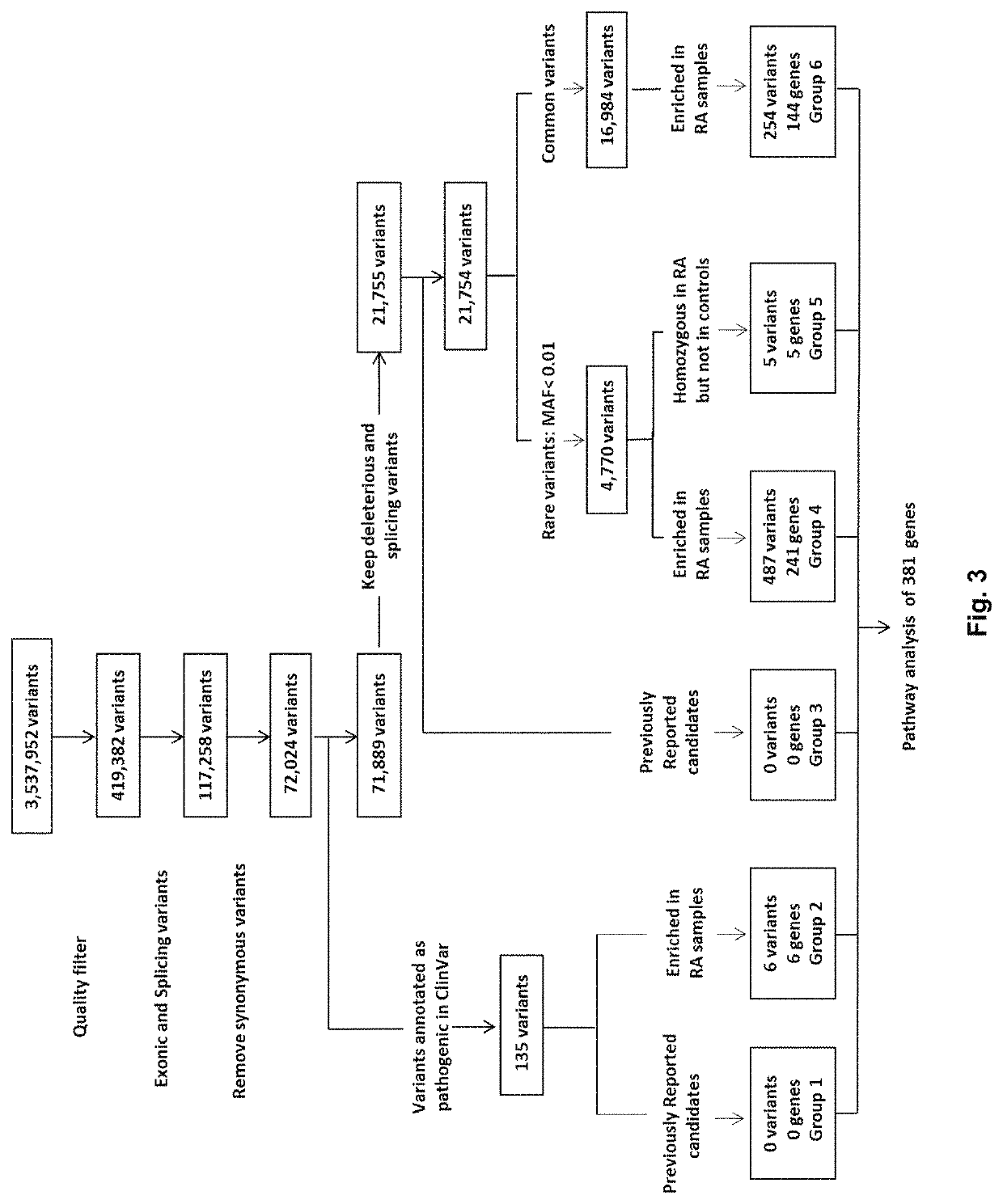
Method of identifying a gene associated with a disease or pathological condition of the disease
a technology of pathological condition and gene, applied in the field of method of identifying a gene associated with a disease, can solve the problems of significant influence, general susceptibleness of studies to genotyping errors, etc., and achieve the effect of improving the genotype accuracy of results and separating rare variants
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Sample Collections
1. Patients
[0048]58 patients diagnosed as having RA were unrelated individuals of Han Chinese descent recruited from hospitals in Southern and Eastern China (Guangzhou and Changzhou) using 2010 Rheumatoid Arthritis Classification Criteria established by American College of Rheumatology and European League Against Rheumatism Collaborative Initiative (2010 ACR / EULAR).
[0049]In addition, 66 healthy and unrelated blood donors of Han Chinese ancestry from Medical Center for Physical Examination and Health Assessment, were included as controls.
[0050]Detailed descriptions of sequenced individuals and clinical characteristics of the enrolled patients are provided in Table 1 and 2. Written informed consent was obtained from all of the participants, and the study was registered in Chinese Clinical Trial Registry (ChiCTR-ROC-17010351) and approved by the local ethics committees of Macau University of Science and Technology (Macau, China).
[0051]
TABLE 1Basic information of the 1...
example 2
Preparation of a List of Candidate Genes Associated with Rheumatoid Arthritis
[0056]A list of 159 candidate RA-associated genetic variants reported by previous genome wide association studies (GWAS) with the P value threshold of P−5, as shown in Table 3, was prepared based on Rheumatoid Arthritis associated genes in the NHGRI GWAS Catalog (Welter D et al., Nucleic acids research 2014; 42:D1001-D1006) and literatures (Freudenberg J et al., Arthritis Rheumatol 2014; 66:1121-1132; Manolio T A et al., Nature 2009; 461:747-753; Okada Y et al., Nature 2014; 506:376-381; and Diogo D et al., The American Journal of Human Genetics 2013; 92:15-27).
[0057]
TABLE 3A list of candidate genes having high priority in RAGeneSNPp-ValueOdd ratioReferencesABHD6rs730815545.00E−081.18Okada Y, PMID: 24390342ACOXLrs67325653.00E−081.07Okada Y, PMID: 24390342AFF3rs9653442| 1.00E−14|1.12|1.12|Okada Y, PMID: 24390342|Stahlrs11676922| 1.00E−14|1.12E A, PMID: 20453842|Jiangrs10865035 2.00E−08|L, PMID: 24782177|Stah...
example 3
Analysis of the Whole-Exome Sequencing (WES)
[0058]To analyze the entire cohort of samples for genotype calls, variant analysis and joint genotyping were performed according to the pipeline recommended by the Genome Analysis Toolkit software and the GATK Best Practices procedures on RA patients and healthy controls (San Lucas F A et al., Bioinformatics 2012; 28:421-422; and Dong C et al., Human molecular genetics 2015; 24:2125-2137). Briefly, Burrows-Wheeler Aligner (BWA) software was utilized to align the raw sequencing reads in FASTQ formats to the 1000 Genomes (GRCh37+decoy) human genome reference. The BWA alignment files were converted to BAM files with SAMtools v1.1, which was used for sorting the BAM files. Duplicate reads were marked for BAM files with Picard MarkDuplicates (https: / / sourceforge.net / projects / picard / ). The coverage and depth were computed based on the final BAM file. Local realignment, base quality recalibration, variant calling, joint genotyping, and variant qu...
PUM
| Property | Measurement | Unit |
|---|---|---|
| truth sensitivity threshold | aaaaa | aaaaa |
| ancestry composition analysis | aaaaa | aaaaa |
| read depth | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com