Viral nucleotide sequences
a nucleotide sequence and virus technology, applied in the field of virus nucleotide sequences, can solve the problems of retained pathogenicity, infectious laryngotracheitis is also a worldwide problem,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
gB Gene of MDV
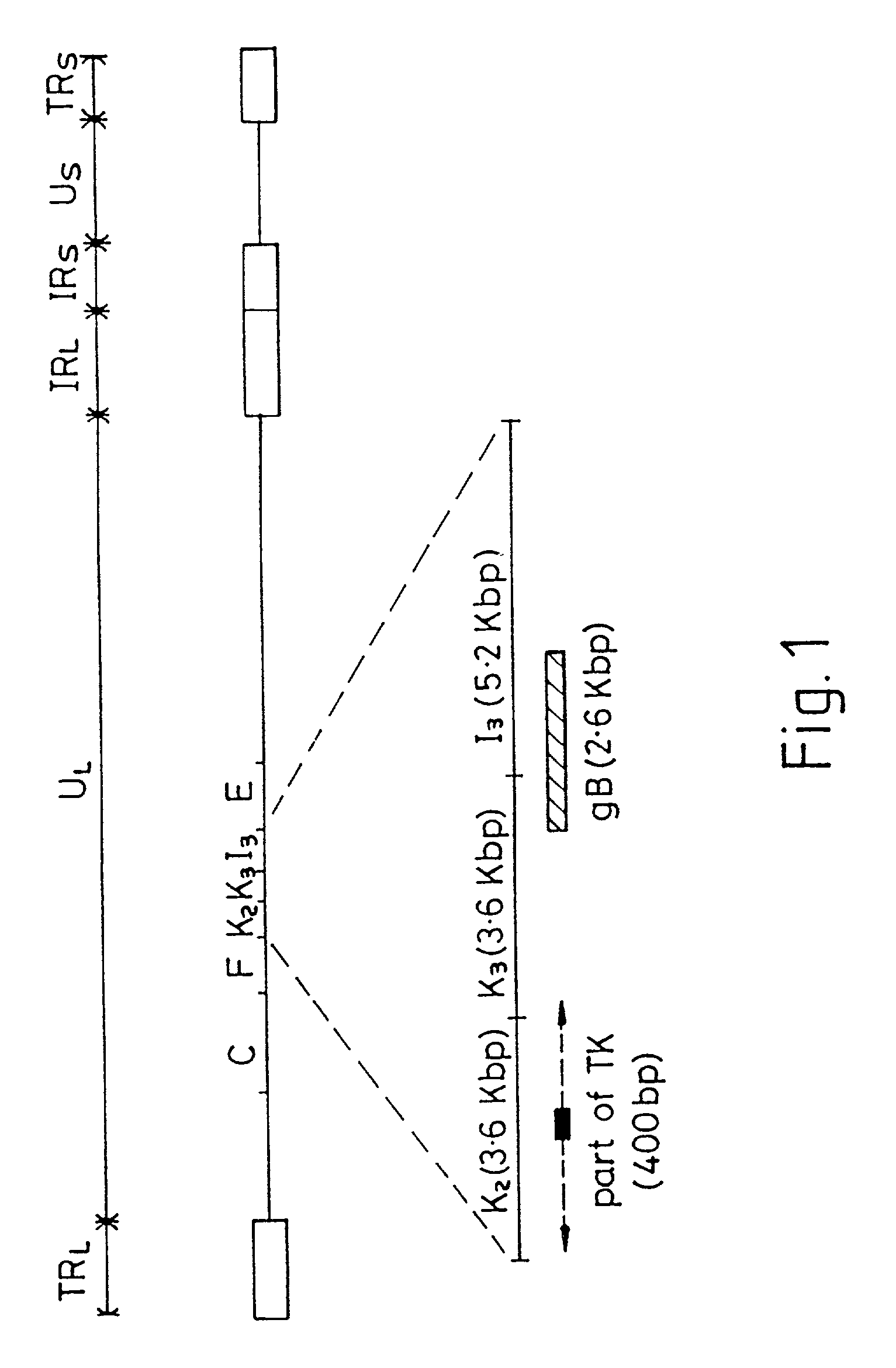
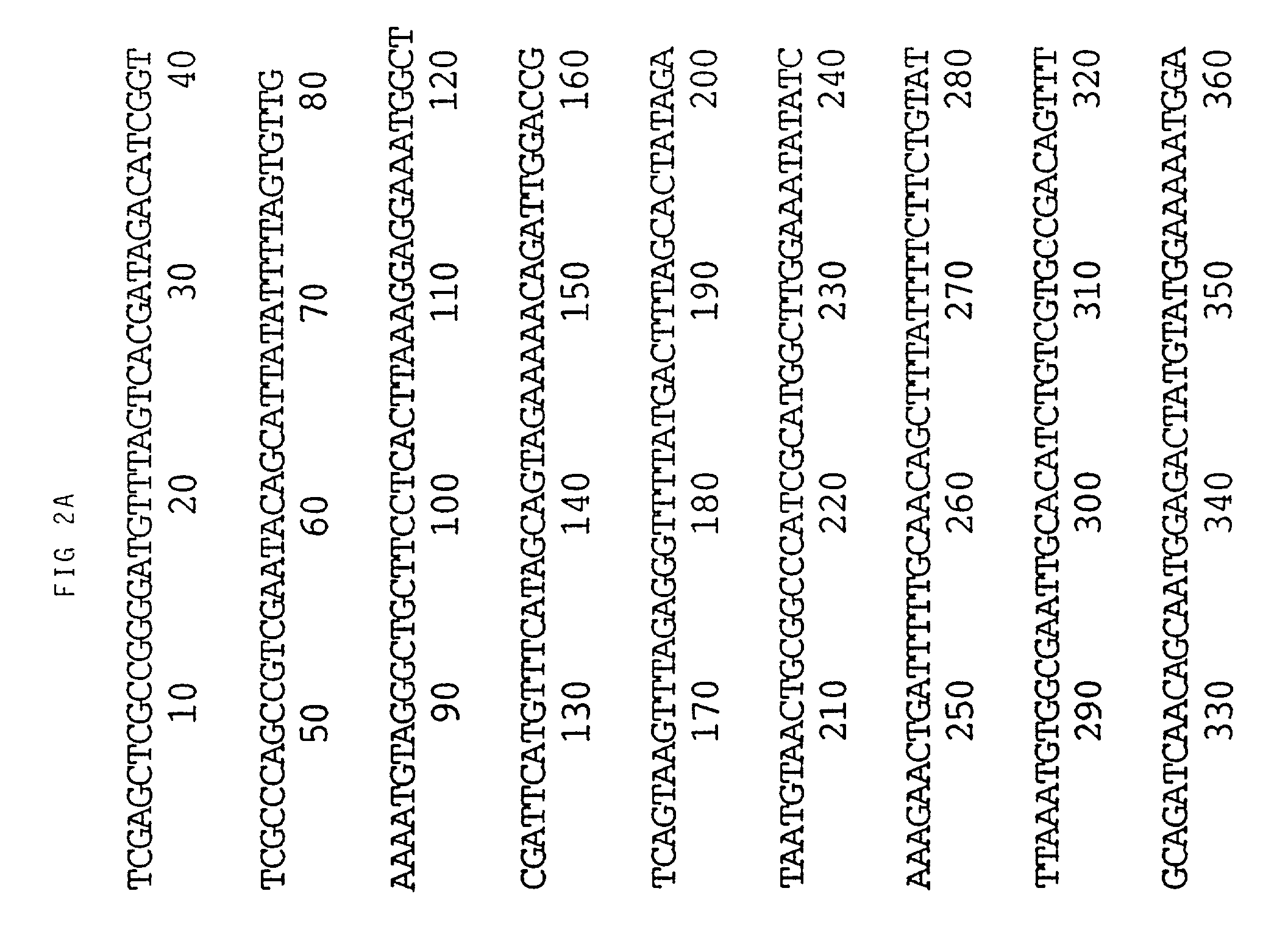
[0095] An M13 clone of HVT homologous to the gB gene of VZV and HSV hybridized to BamH1 fragment I3 of MDV (see FIG. 1). Sequencing of this fragment obtained from a BamH1 library of the RB1B strain of MDV showed that two thirds of the gene, starting with the NH.sub.2 terminus, was contained within I3. The remainder of the gene was identified in the adjacent restriction fragment K3. FIG. 1 shows the map position of the gene which is 2.6 Kbp long. Its mRNA has been estimated to be approximately 2.8 Kb. The translated protein is 865 amino acids long (FIG. 2). This includes approximately 20 amino acids which may be part of a signal sequence domain. The primary translated sequence of MDV gB has a few features in common with gB of other herpes viruses such as the alignment of cysteine residues and the presence of hydrophobic sequences which are presumably capable of spanning a lipid bilayer [Pellet, P. E. et al (1985), J. Virol. 53, 243-253]. However, MDV gB has only 48% ami...
example 2
gH Gene of HVT and gH Gene of MDV
[0096] An M13 clone of HVT containing sequences homologous to HSV gH was isolated during our earlier work on gene identification and mapping (Buckmaster et al (1988) as above). This clone, when used as a probe, hybridized to a 6 Kbp HindIII fragment of HVT (FIG. 3). Sequencing revealed that this fragment contained approximately one quarter of the gH gene including the carboxy terminus. The adjacent HindIII fragment (3.2 Kbp) containing the remainder of the gH gene was identified by hybridization using a cloned HpaI fragment of HVT which overlapped the HindIII site. FIG. 4 shows the sequence of the coding region of the gH gene of HVT (2.3 Kbp) and flanking sequences. The % amino acid identity between the gH gene of HVT and its homologue in HSV1, VZV and EBV was only 20, 24 and 20 respectively (estimated from maximised amino acid overlaps of 630, 644 and 153 respectively).
example 3
TK Gene of HVT and TK Gene of MDV
[0097] The whole coding region of the TK gene of HVT (1053 bp) was contained within the 3.2 Kbp HindIII fragment described above (FIG. 3). The sequence of the entire gene and flanking regions is shown in FIG. 5. Similarly the whole of the MDV TK gene is contained within the 3.6 Kbp BamH1 K2 fragment of MDV (FIG. 1). The sequence of MDV TK gene determined so far is shown in FIG. 5. Comparison of the MDV and HVT TK sequences indicates that the two genes have approximately 60% amino acid identity (estimated from 276 amino acid overlap). By contrast, the % amino acid identities between the TK gene of HVT and the TK genes of HSV 1, VZV and EBV are only 30, 27 and 24 respectively (estimated from amino acid overlaps of 320, 332 and 193 respectively). The predicted amino acid sequences of HVT and MDV TK show characteristic ATP and / or CTP binding site motifs described for a number of virus and eukaryotic proteins that are associated with phosphorylation (Gent...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| genome structure | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com