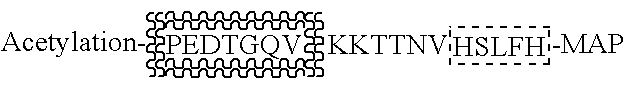Methods for reducing complexity of a sample using small epitope antibodies
a technology of epitope antibodies and small sample sizes, applied in the field of methods for can solve the problems of large sample sizes, time-consuming, and limited in the ability to reproduce a significant fraction, and achieve the effect of reducing the complexity of samples
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Preparation and Characterization of Small Epitope Antibodies
[0198] Five immunization polypeptides in the format of Multiple Antigenic Peptide (MAP) were designed as shown in Table 3. These sequences in combination were also used to evaluate cross-reactivity of the induced antibodies, by virtue of the inclusion in different MAPs of the same sequence in differing locations. Each of the immunization polypeptides was used to immunize 4 Balb / C mice using standard methods.
TABLE 3Design of immunization polypeptidesPeptideGroupSequenceSEQ ID NOMAP11MAP22MAP33MAP44MAP55
Notes to Table 3:
Polypeptide MAP1: HSLFHPEDTGQV: From PSA, amino acids #79-89. KKTTNV: From Meningococcal Opa protein, containing KTT, a published 3mer antibody epitope (Malorny, Morelli et al. 1998).
Polypeptide MAP2: Alternate sequences of MAP 1.
Polypeptide MAP3: LTPKK: Motif 1 of PSA (Nagasaki, Watanabe et al. 1999). KKTTNVLTVPTNIPG: From Meningococcal Opa protein, containing two published 3mer antibody epitopes: KTT a...
example 2
Preparation of Small Epitope Antibodies
[0213] An approach to identify antibodies based on phage display antibody screening was performed. Five peptide sequences used for the selection of positive antibodies are shown in Table 9. These sequences in combination were also used to evaluate cross-reactivity of the selected antibodies.
TABLE 9Design of screening polypeptidesPeptideSequenceP1CXXXXXDTGXXXXXXP6CXXXXXDTGXXXXXXP7CXXXXXAQVXXXXXXP8CXXXXXIARXXXXXXP9CXXXXXLSHXXXXXX
[0214] Note to Table 9: The letter ‘X’ denotes a mixture of the naturally-occurring L-amino acids excluding cysteine, methionine, and tryptophan.
[0215] Positives were selected after six rounds of enrichment. The results of phage ELISA screens against the five screening peptides is shown in Table 10. A total of 96 phage were screened for P1; 48 were screened for polypeptides P6-P9. In all cases, positive phage were identified above background.
TABLE 10Reactivity of enriched phage against screening polypeptidesPolypept...
example 3
Protein Profiling and Biomarker Development
[0218] In one exemplary method for protein profiling, serums derived from healthy and affected individuals for a particular disease of clinical interest are subjected to: (a) debulking of the most abundant protein constituents; (b) deglycosylation of the less abundant proteins that remain; (c) reduction and alkylation of cysteine residues present in the debulked proteome; (d) digestion of the debulked proteome to completion; (e) fractionation of the resulting peptide fragments with small epitope antibodies as described above; and (f) comparison of the composition and relative abundance of peptide constituents from epitope enriched fractions derived from healthy and affected patients to identify candidate biomarkers associated with a specific disease.
[0219] Fractionation with small epitope antibodies is performed in parallel with a set of approximately 100 small epitope antibodies of different specificities. Each antibody is chosen based o...
PUM
| Property | Measurement | Unit |
|---|---|---|
| chemical | aaaaa | aaaaa |
| mass spectrometry | aaaaa | aaaaa |
| structure | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com