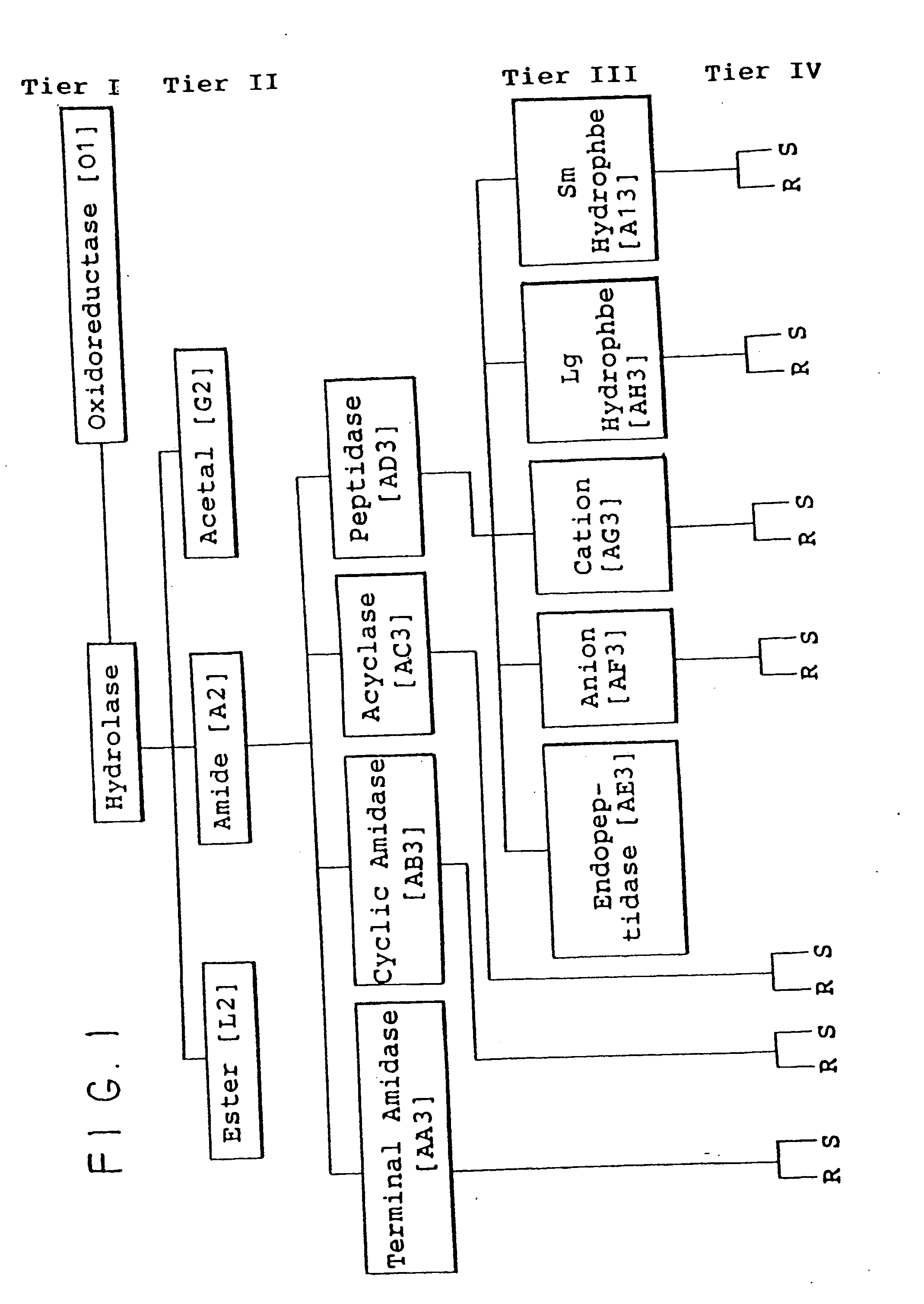
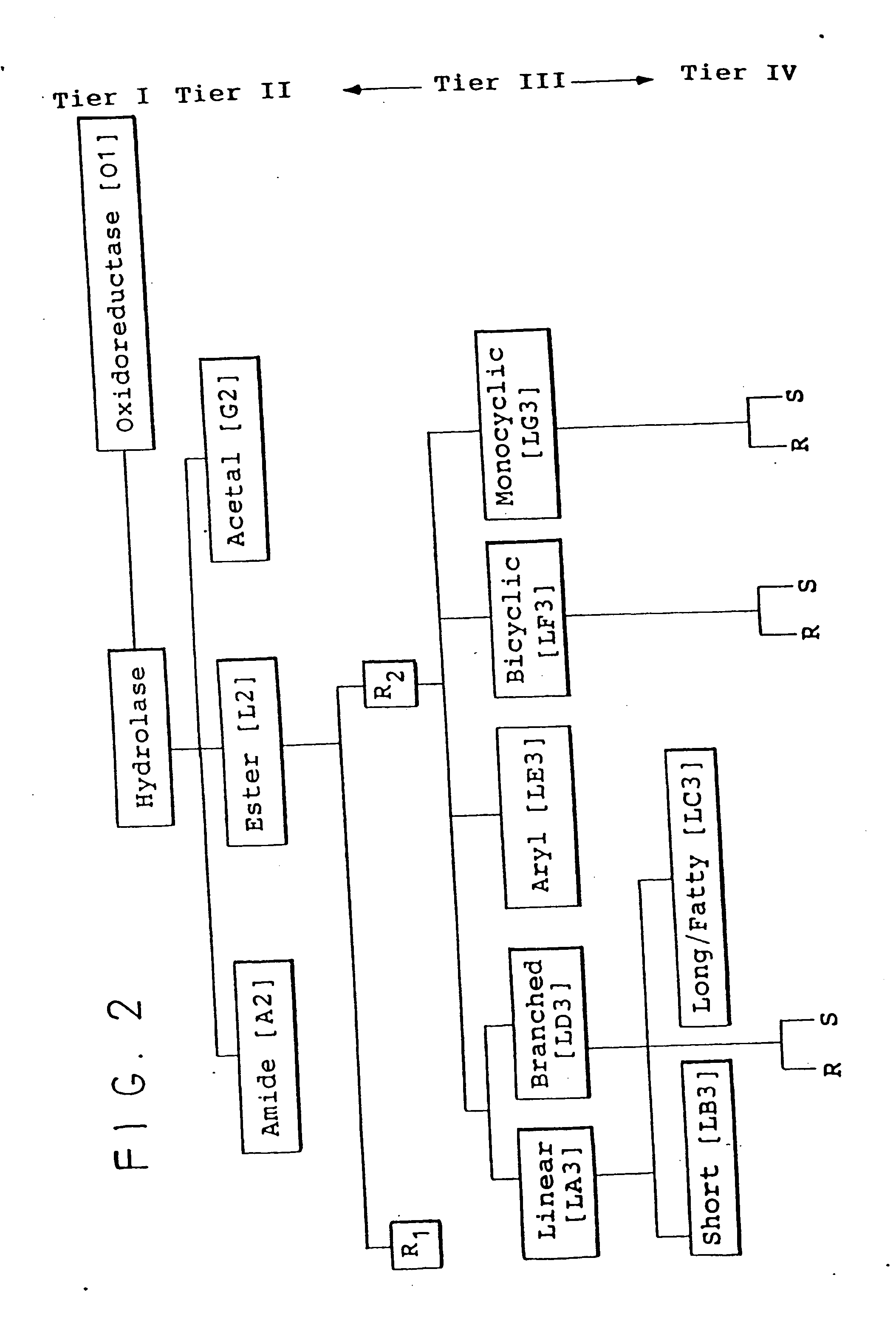
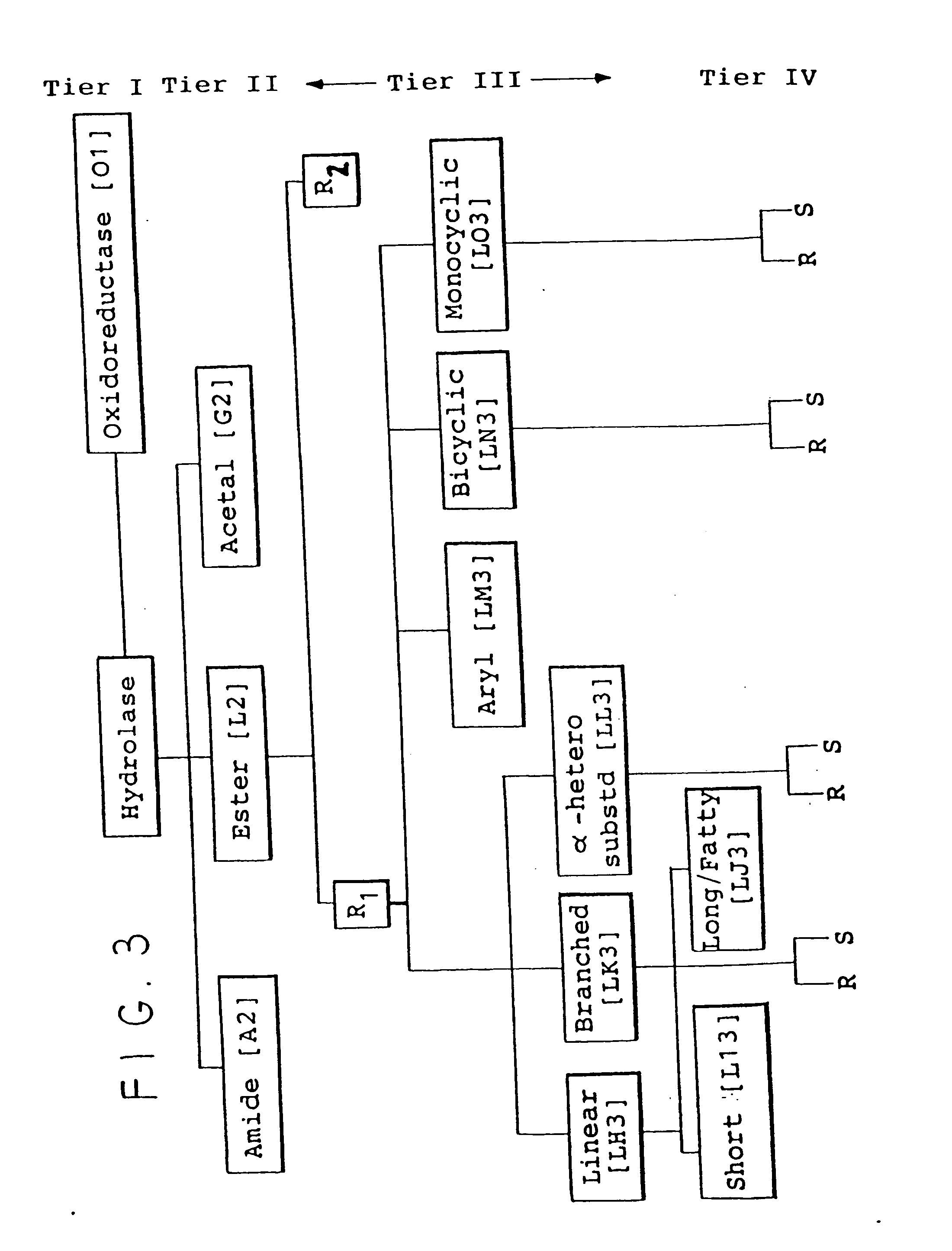
Protein activity screening of clones having DNA from uncultivated microorganisms
a technology of protein activity and clone, which is applied in the field of preparing and screening libraries of clones containing microbially derived dna, can solve the problems of large fraction of this diversity so far unrecognized, difficult or impossible to identify or isolate valuable proteins, e.g. enzymes, from these samples, and achieves stably integrating large segments of genomic dna. , the effect of less transfer
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 3
Construction of a Stable, Large Insert Picoplankton Genomic DNA Library
[0291]FIG. 5 shows an overview of the procedures used to construct an environmental library from a mixed picoplankton sample. A stable, large insert DNA library representing picoplankton genomic DNA was prepared as follows.
[0292] Cell collection and preparation of DNA. Agarose plugs containing concentrated picoplankton cells were prepared from samples collected on an oceanographic cruise from Newport, Oreg. to Honolulu, Hi. Seawater (30 liters) was collected in Niskin bottles, screened through 10 μm Nitex, and concentrated by hollow fiber filtration (Amicon DC10) through 30,000 MW cutoff polyfulfone filters. The concentrated bacterioplankton cells were collected on a 0.22 1 lm, 47 mm Durapore filter, and resuspended in 1 ml of 2× STE buffer (1M NaCI, 0.1M EDTA, 10 mM Tris, pH 8.0) to a final density of approximately 1×1010 cells per ml. The cell suspension was mixed with one volume of 1% molten Seaplaque LMP ag...
PUM
| Property | Measurement | Unit |
|---|---|---|
| depth | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com