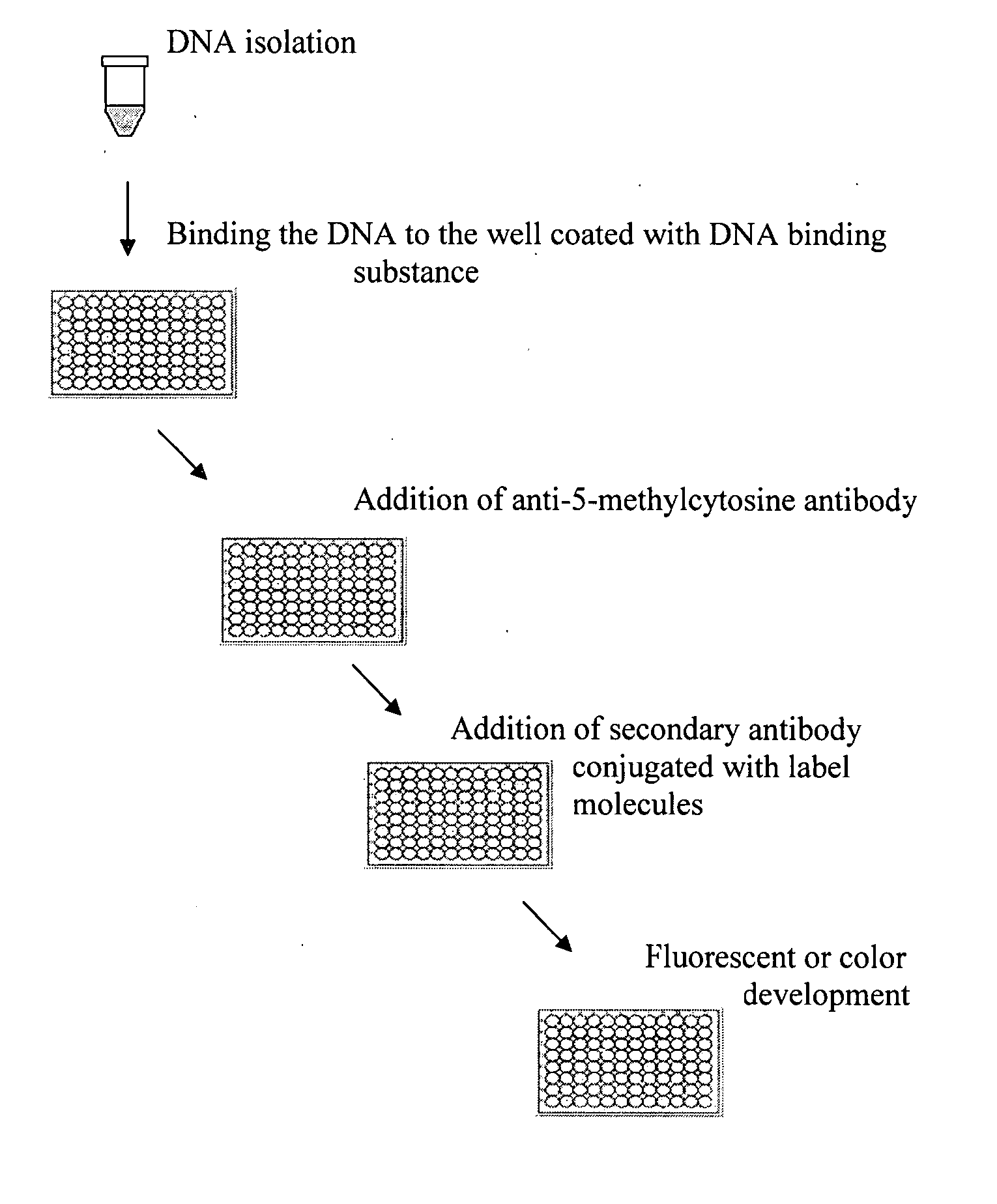
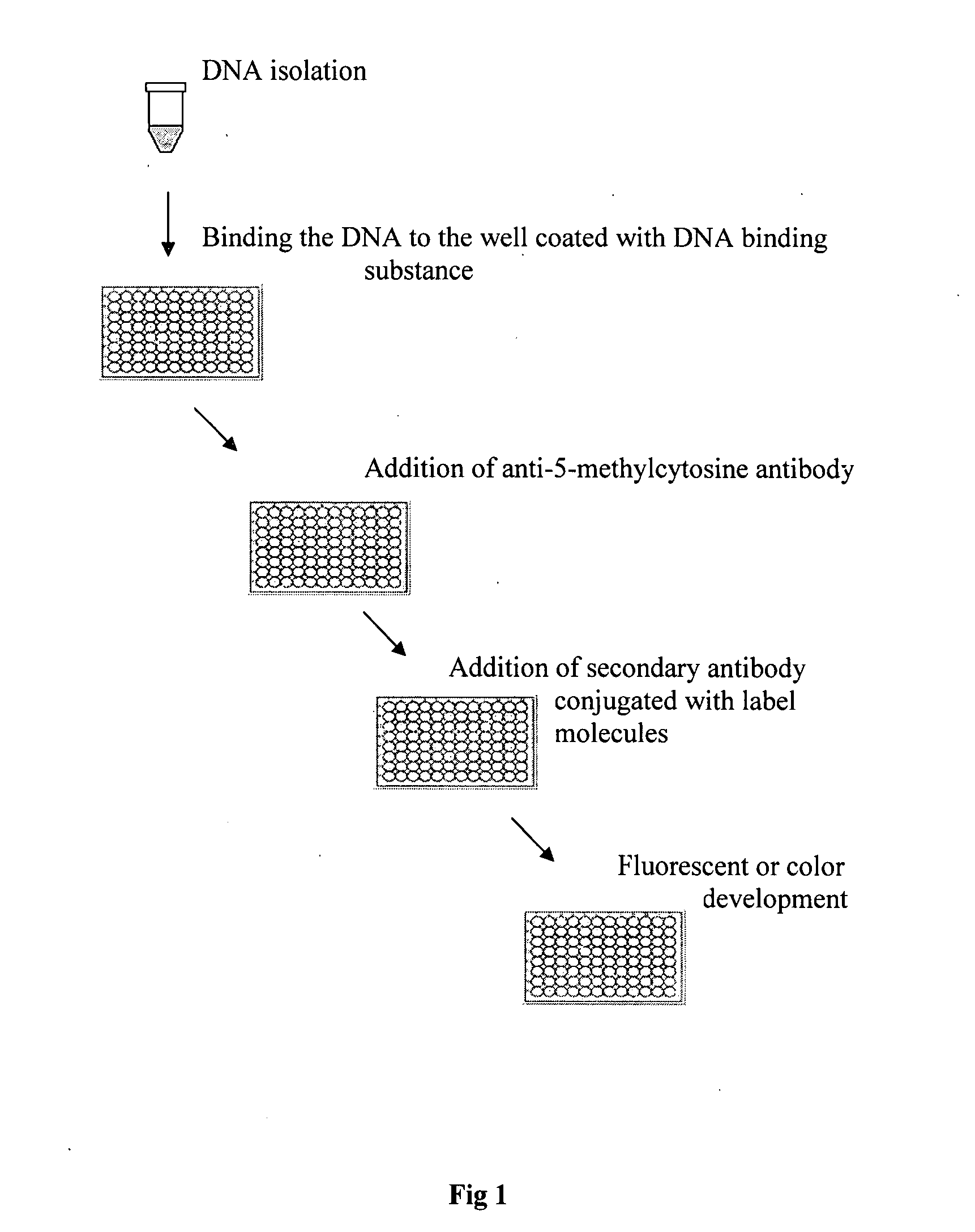
Method of rapidly quantifying global DNA methylation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
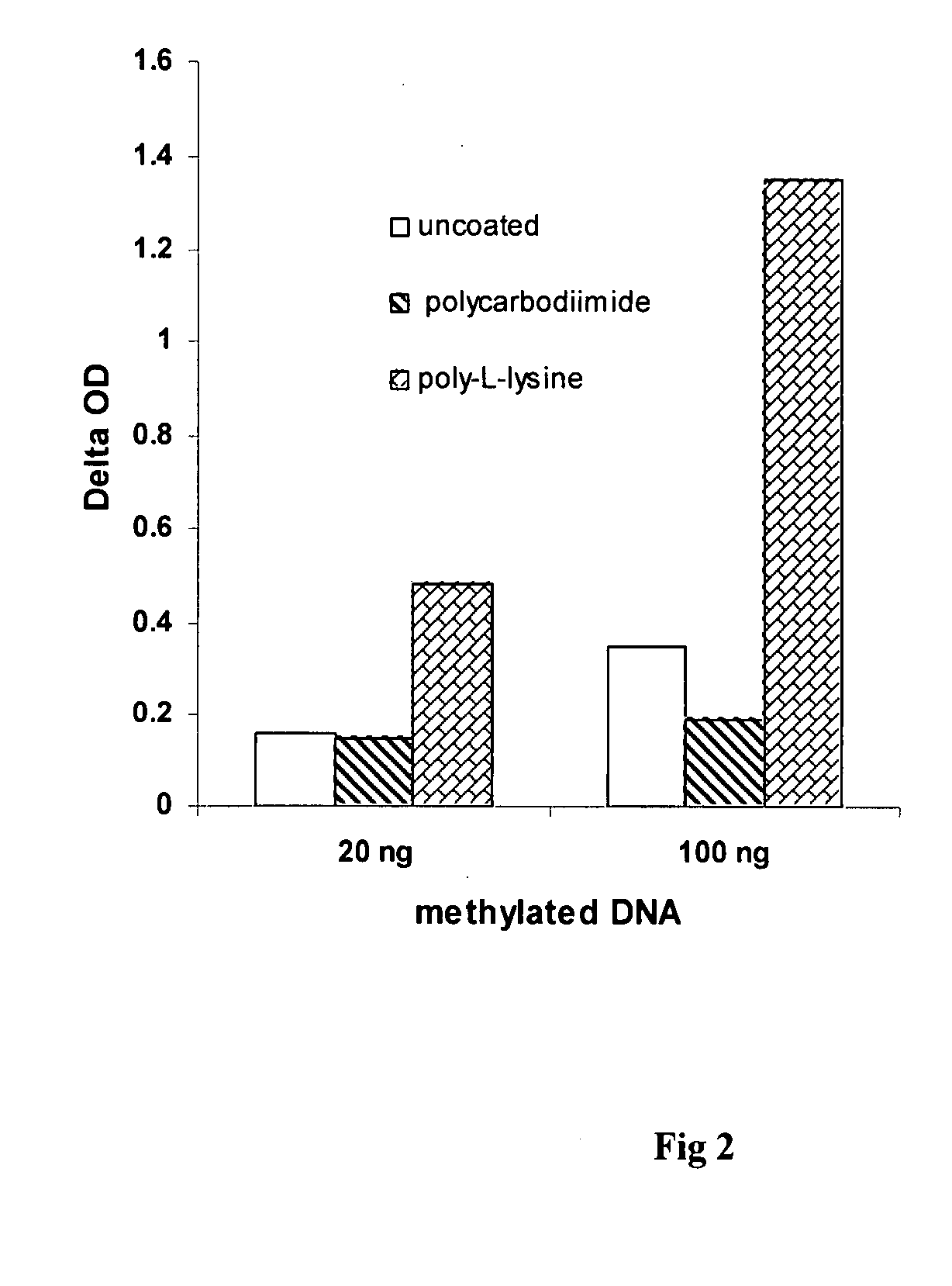
[0030]The experiment was carried out to compare the effect of different nucleic acid binding substance coating on the detection of global DNA methylation. Polystyrene 8-well strips were coated with 0.01% poly-L-lysine, or 1% polycarbodiimide, respectively. After storage at 4° C. for 2 weeks, the coated and uncoated strips were used for DNA immobilization. 30 μl of naturally methylated NY-2A virus DNA at different concentrations were added into the strip wells. The strips were then incubated at 37° C. for 1.5 h followed by incubation at 60° C. for 30 min to dry the wells. The wells were blocked with 2% BSA solution at 37° C. for 30 min followed by washing 3 times. 50 μl of monoclonal anti-5-methylcytosine antibody was then added at 0.5 pg / ml and incubated at room temperature for 1 h. The wells were washed 3 times with PBS containing 0.1% tween-20 after the antibody solution was removed. 50 μl of anti-mouse antibody conjugated with HRP at 0.1 μg / ml was added into the wells and incubat...
example 2
[0031]The experiment was carried out to compare the effect of dry-capture method and conventional immobilization methods on the detection of global DNA methylation. Polystyrene 8-well strips were coated with 0.01% poly-L-lysine. After storage at 4° C. for 2 weeks, the coated strips were used for DNA immobilization. With the method of this invention, naturally methylated NY-2A virus DNA was diluted with 1× TE buffer. 30 μl of diluted virus DNA at different concentrations were added into the strip wells. The strips were then incubated at 37° C. for 1.5 h followed by incubation at 60° C. for 30 min to dry the wells. With the conventional method, 50 μl of diluted virus DNA at different concentrations were added into the wells and incubated at room temperature for 4 h or at 4° C. overnight. The remaining solution was removed after incubation. The DNA-immobilized wells with both methods were blocked with 2% BSA solution at 37° C. for 30 min followed by washing 3 times. 50 μl of monoclonal...
example 3
[0032]The experiment was carried out to examine the detection sensitivity of the method based on this invention on the quantification of global DNA methylation. Polystyrene 8-well strips were coated with 0.01% poly-L-lysine. After storage at 4° C. for 2 weeks, the coated strips were used for DNA immobilization. Naturally methylated NY-2A virus DNA was diluted with 1× TE buffer to different concentrations. 30 μl of diluted virus DNA were added into the wells at the amounts of 2 ng-100 ng / well. The strips were then incubated at 37° C. for 1.5 h followed by incubation at 60° C. for 30 min to dry the wells. The wells were then blocked with 2% BSA solution at 37° C. for 30 min followed by washing 3 times. 50 μl of monoclonal anti-5-methylcytosine antibody was then added at 0.5 μg / ml and incubated at room temperature for 1 h. The wells were washed 3 times with PBS containing 0.1% tween-20 after the antibody solution was removed. 50 μl of anti-mouse antibody conjugated with HRP at 0.1 μg / m...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Temperature | aaaaa | aaaaa |
| Temperature | aaaaa | aaaaa |
| Temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com