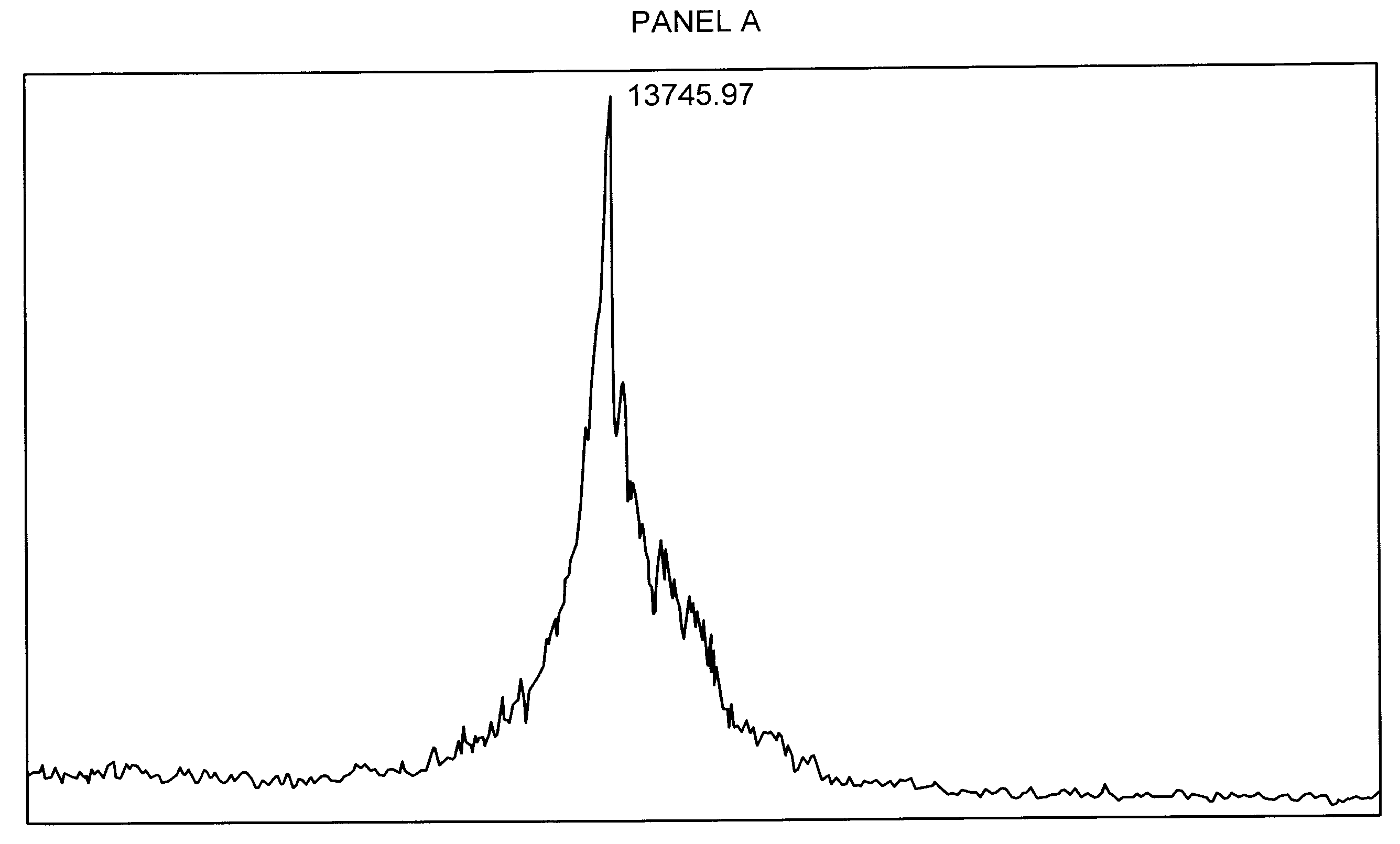
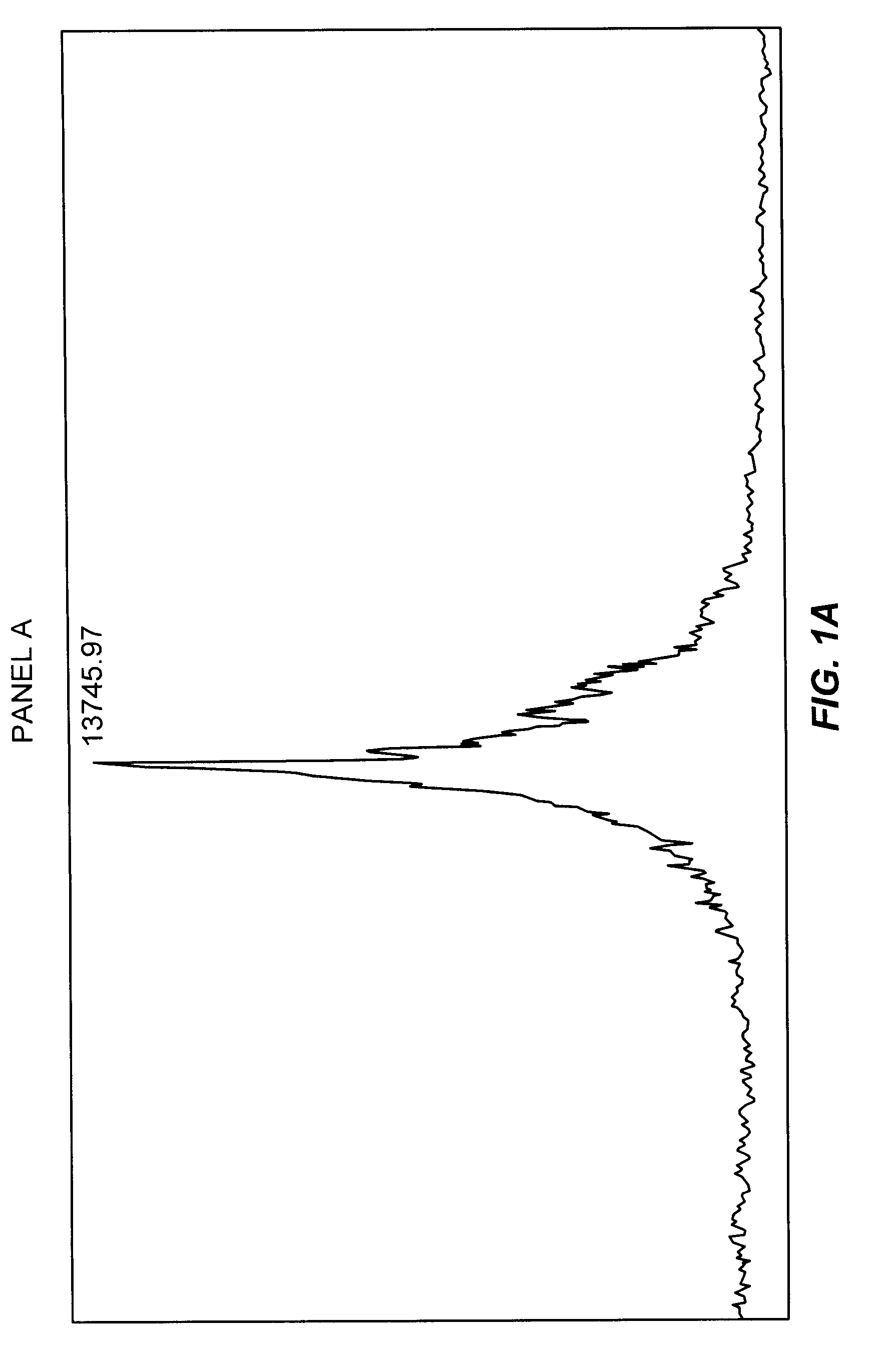
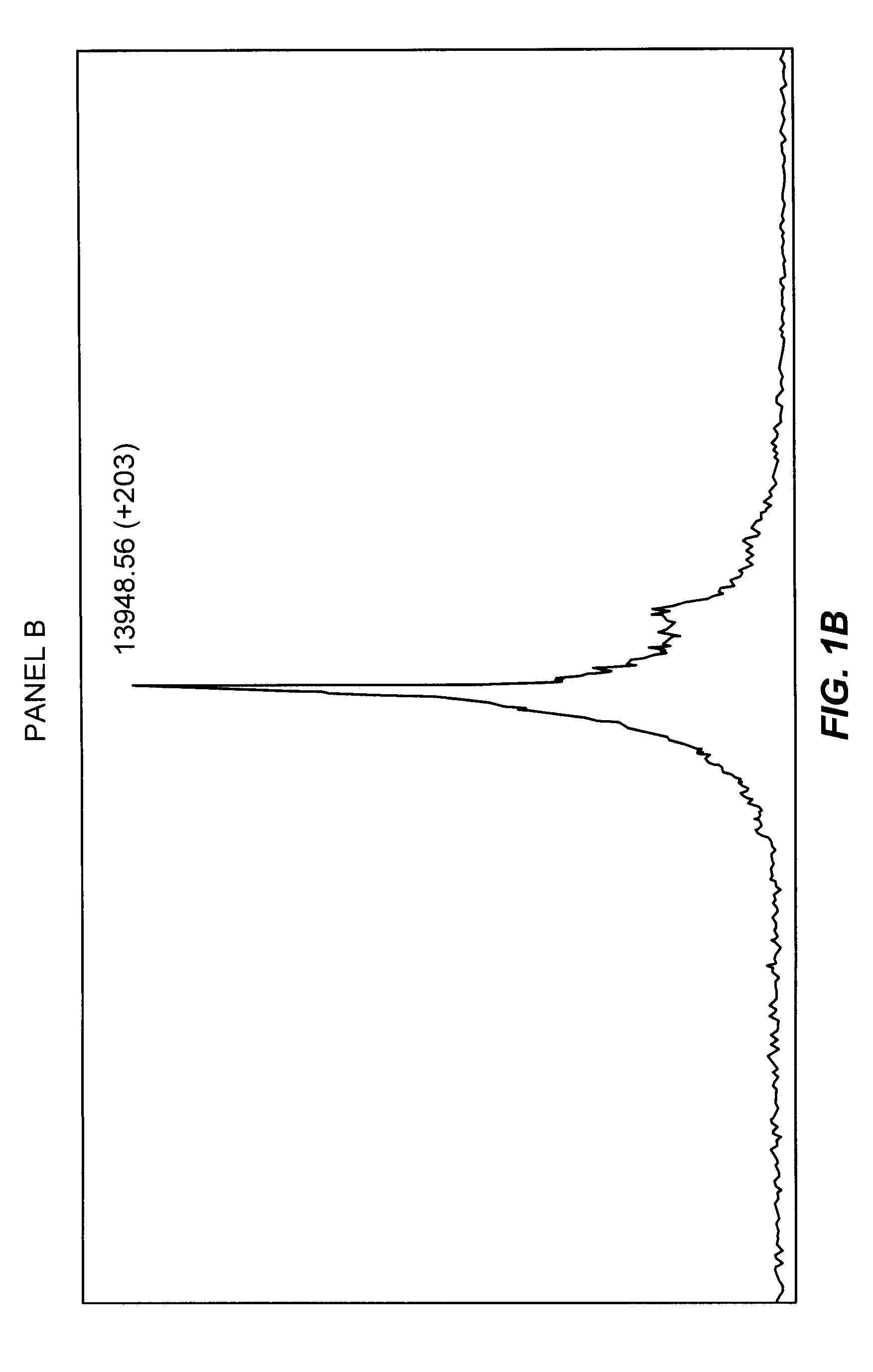
Glycosylation of peptides via o-linked glycosylation sequences
a glycosylation sequence and peptide technology, applied in the field of polypeptide modification by glycosylation, can solve the problems of rapid neutralization of peptides and/or allergic reactions, limited use of such polypeptides as therapeutic agents, and lack of homogeneity of the final produ
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Incorporation of Glycosylation Sites into Bone Morphogenetic Protein-7 (BMP-7)
1.1. BMP-7 Sequence Information
[0709]An exemplary BMP-7 sequence is shown below (S.1).
Human Bone morphogenetic protein-7(SEQ ID NO: 164)M1STGSKQRSQNRSKTPKNQEALRMANVAENSSSDQRQACKKHELYVSFRDLGWQDWIIAPEGYAAYYCEGECAFPLNSYMNATNHAIVQTLVHFINPETVPKPCCAPTQLNAISVLYFDDSSNVILKKYRNMVVRACGCH
[0710]The N-terminal methionine may be present or absent in any BMP-7 mutant. In this example, the numbering of the amino acid residues is based on the initial unmodified sequence in which the left most residue, methionine (M), is numbered as position 1. To highlight how the mutant sequence differs in respect to the unmodified sequence, the numbering of unmodified amino acids as they appear in the mutant sequences below remains unchanged following the modification. More than one of the described sequence modifications may be present in a BMP-7 mutant of the present invention.
[0711]Preferred regions for introduction of mutations to cre...
example 2
Incorporation of Glycosylation Sequences into Neutrotrophin-3 (NT-3)
2.1. NT-3 Sequence Information
[0725]An exemplary wild-type amino acid sequence (S.2) of human NT-3 is shown below.
Human Neurotrophin-3 (SEQ ID NO: 340):
[0726]MYAEHKSHRGEYSVCDSESLWVTDKSSAIDIRGHQVTVLGEIKTGNSPVKQYFYETR CKEARPVKNGCRGIDDKHWNSQCKTSQTYVRALTSENNKLVGWRWIRIDTSCVCAL SRKIGRT
[0727]This example describes amino acid sequence mutations introducing O-linked glycosylation sequences into the wild-type NT-3 sequence S.2 (SEQ ID NO: 340) or any modified (e.g., previously mutated) version thereof. A number of mutants were created introducing O-linked glycosylation sites into 3 loop regions as well as the amino terminus.
[0728]The N-terminal methionine (M) may be present or absent in any NT-3 mutant. In this example, the numbering of the amino acid residues is based on the initial unmodified sequence in which the N-terminal residue, methionine (M), is numbered as position 1. To highlight how the mutant sequence differs wit...
example 3
Expression of Human BMP-7 and Human NT-3 Using Various Vectors and E. coli Host Cells
[0745]The BMP-7 native sequence S.1 (SEQ ID NO: 164) and the above described BMP-7 mutants C.1 to C.31 (SEQ ID NOs: 273-277, 279-283, 285-287, 289-291, 293-296, 298-305, 307-309) (Example 1) as well as the NT-3 native sequence S.2 (SEQ ID NO: 340) and the above described NT-3 mutants A.1-A.16 (SEQ ID NOs: 342, 343, 345-347, 349-351, 353-360) (Example 2) can be expressed using a variety of vectors in different E. coli host cells. Experimental results for the native sequences are summarized in Table 17, below. In addition, all BMP-7 mutants C.1 to C.31 were expressed in W3110 E. coli at 37° C. as inclusion bodies.
TABLE 17Expression of native human BMP-7 (S.1) (SEQ ID NO: 164)and native NT-3 (S.2) (SEQ ID NO: 340) in E. coliE. coli HostInductionProteinVectorCellTemperatureBMP-7pET24atrxb,gor,supp-2 DE320° C.BMP-7pET24aNovaBlue(DE3)37° C.BMP-7pET24aNovaBlue(DE3)20° C.BMP-7pcWin2W311037° C.NT-3pET24atrxb...
PUM
| Property | Measurement | Unit |
|---|---|---|
| covalent | aaaaa | aaaaa |
| negative charge | aaaaa | aaaaa |
| nucleic acid | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com