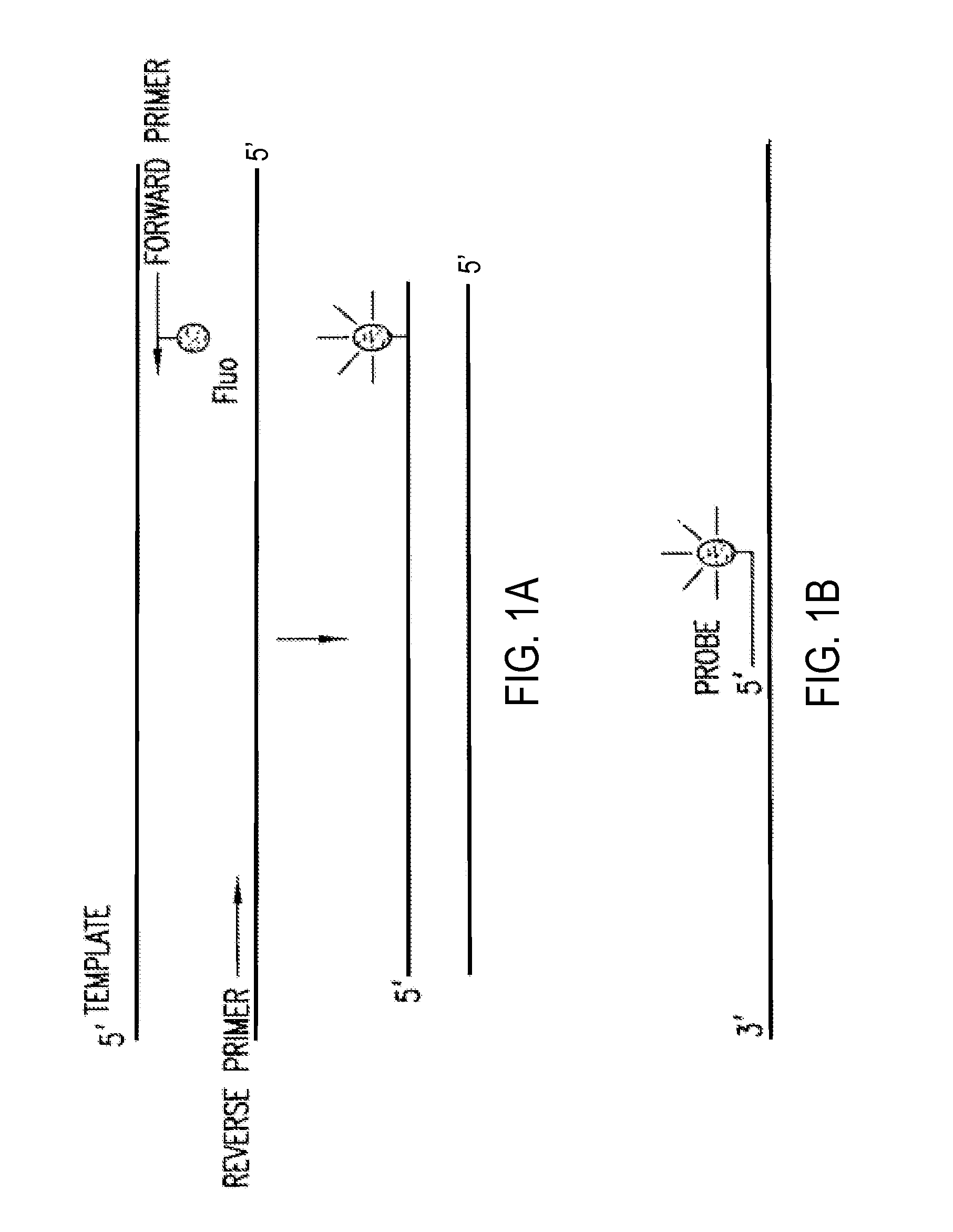
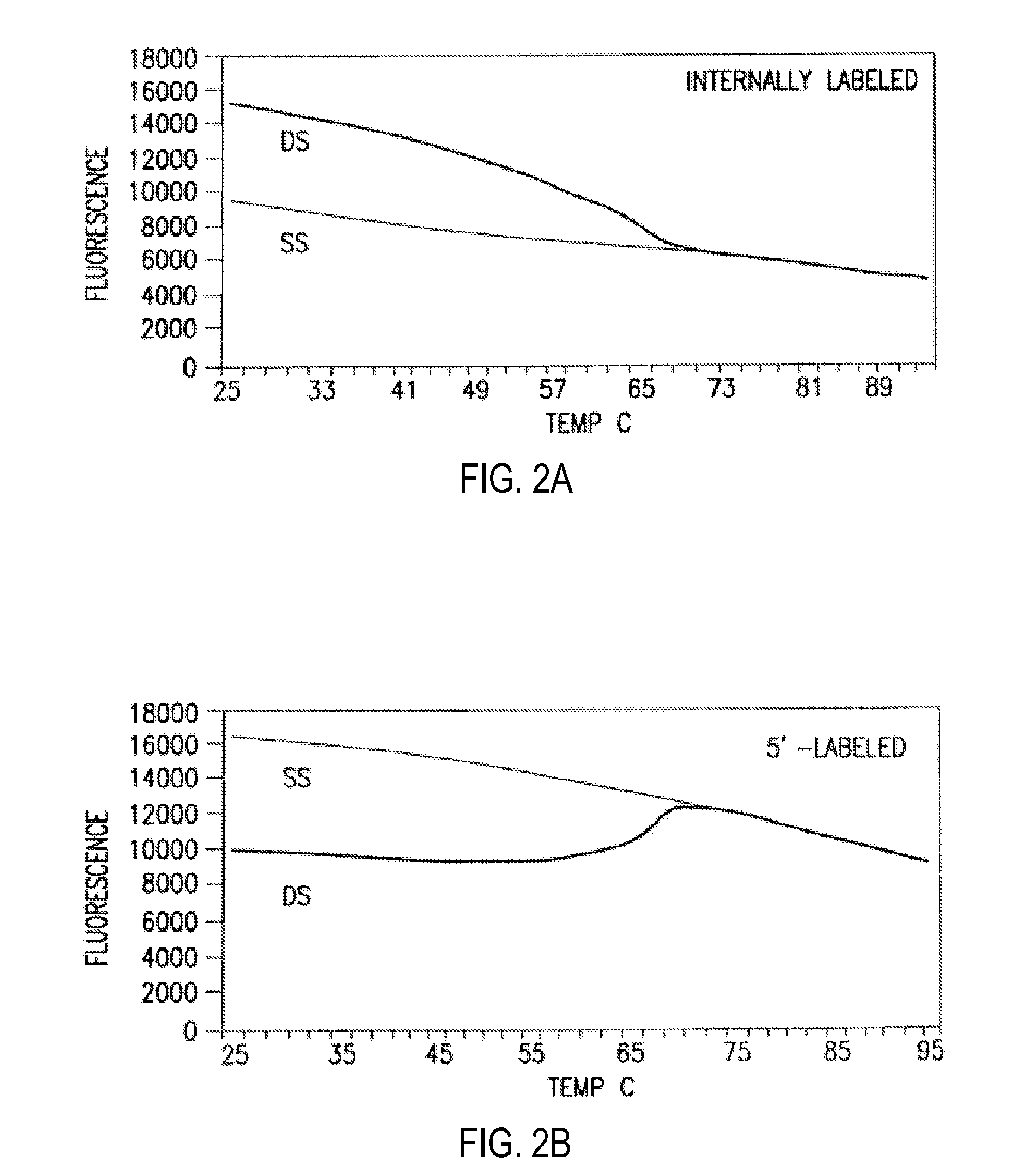
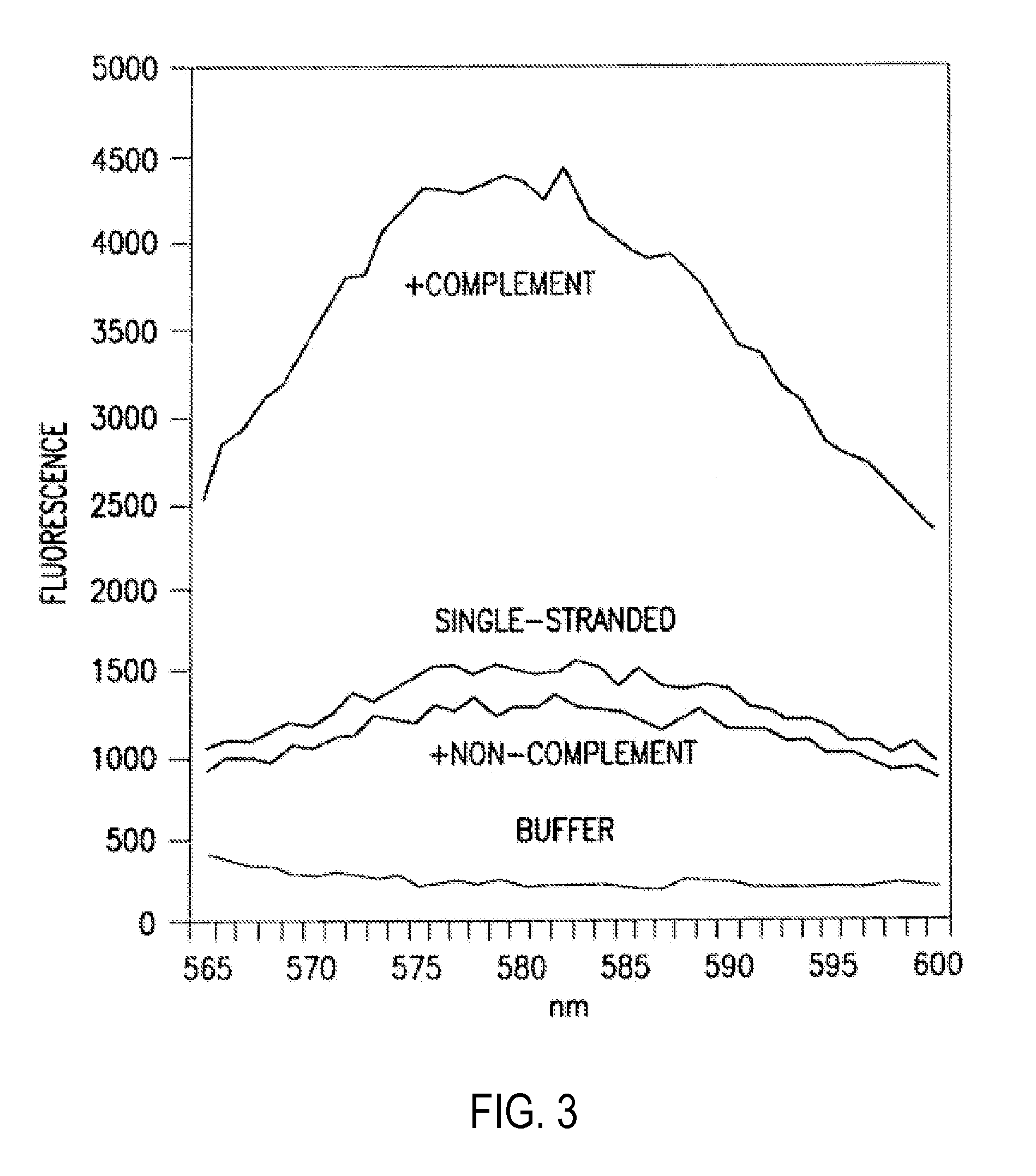
Primers and methods for the detection and discrimination of nucleic acids
a nucleic acid and primer technology, applied in the field of molecular biology, can solve the problems of requiring the sensitivity of such assays remains limited, and the inability to detect nucleic acids in the presence of two modified nucleotides, so as to reduce the background activity of the label, reduce the fluorescence background, and enhance the detection of target nucleic acid molecules
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Preparation of Oligonucleotides
[0173]Oligonucleotides may be prepared using any known methodology. In some preferred embodiments, oligonucleotides may be synthesized on solid supports using commercially available technology. Oligodeoxynucleotides were synthesized using DNA synthesizer-8700 (Milligen / Biosearch). Fluorescent moieties may be incorporated into the oligonucleotides of the present invention using any conventional technology. For example, fluorescent labels may be incorporated into nucleoside phosphoramidites and directly incorporated into the oligodeoxynucleotides during automated chemical synthesis. In some preferred embodiments, the modified nucleotide may be a fluorescein-dT phosphoramidite (Glen Research Cat #10-1056) which may be inserted into designated position during chemical synthesis of oligonucleotide. 5′-fluorescein phosphoramidite (FAM) (Glen Research Cat # 10-5901) and 3′-TAMRA-CPG 500 (Glen Research cat #20-5910) were used to add the indicated labels to the...
example 2
PCR Targets and Conditions
[0183]Those skilled in the art will appreciate that any nucleic acid that can be amplified by PCR may be used in the practice of the present invention.
[0184]Examples of suitable nucleic acids include, but are not limited to, genomic DNAs, cDNAs and cloned PCR products. The practice of the present invention is not limited to use with DNA molecules. For example, mRNA molecules may be used as templates for an amplification reaction by first conducting a first strand synthesis reaction using techniques well known in the art. The present invention has been exemplified using cDNAs for IL4 and b-actin synthesized using total mRNA from the corresponding cells and SuperScript™ System for the First Strand cDNA Synthesis (Gibco BRL, cat #18089-011) according to the manufacturer's manual. IL4 and b-actin cDNAs were amplified and cloned into pTEPA plasmid according to Gibco BRL manual (cat #10156-016).
[0185]The selection of suitable PCR conditions is within the purview ...
example 3
Detection of Nucleic Acids
[0186]Nucleic acids may be detected by any conventional technology. In some preferred embodiments, the nucleic acid to be detected may be a PCR product and may be detected either by agarose gel electrophoresis or by homogeneous fluorescence detection method as described in U.S. provisional patent application Ser. No. 60 / 139,890, filed Jun. 22, 1999. In this method fluorescent signal is generated upon the incorporation of the specifically labeled primer into the PCR product. The method does not require the presence of any specific quenching moiety or detection oligonucleotide. In some preferred embodiments, the detection oligonucleotides are capable of forming a hairpin structure and are labeled with fluorescein attached close to the 3′-end.
[0187]The fluorescent measurements were performed in the PCR reaction buffer using on ABI PRIZM™7700 Sequence Detector, fluorescent plate reader (TECAN) or Kodak EDAS Digital Camera. Excitation / emission wavelengths were 4...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| fluorescent | aaaaa | aaaaa |
| nucleic acid | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com