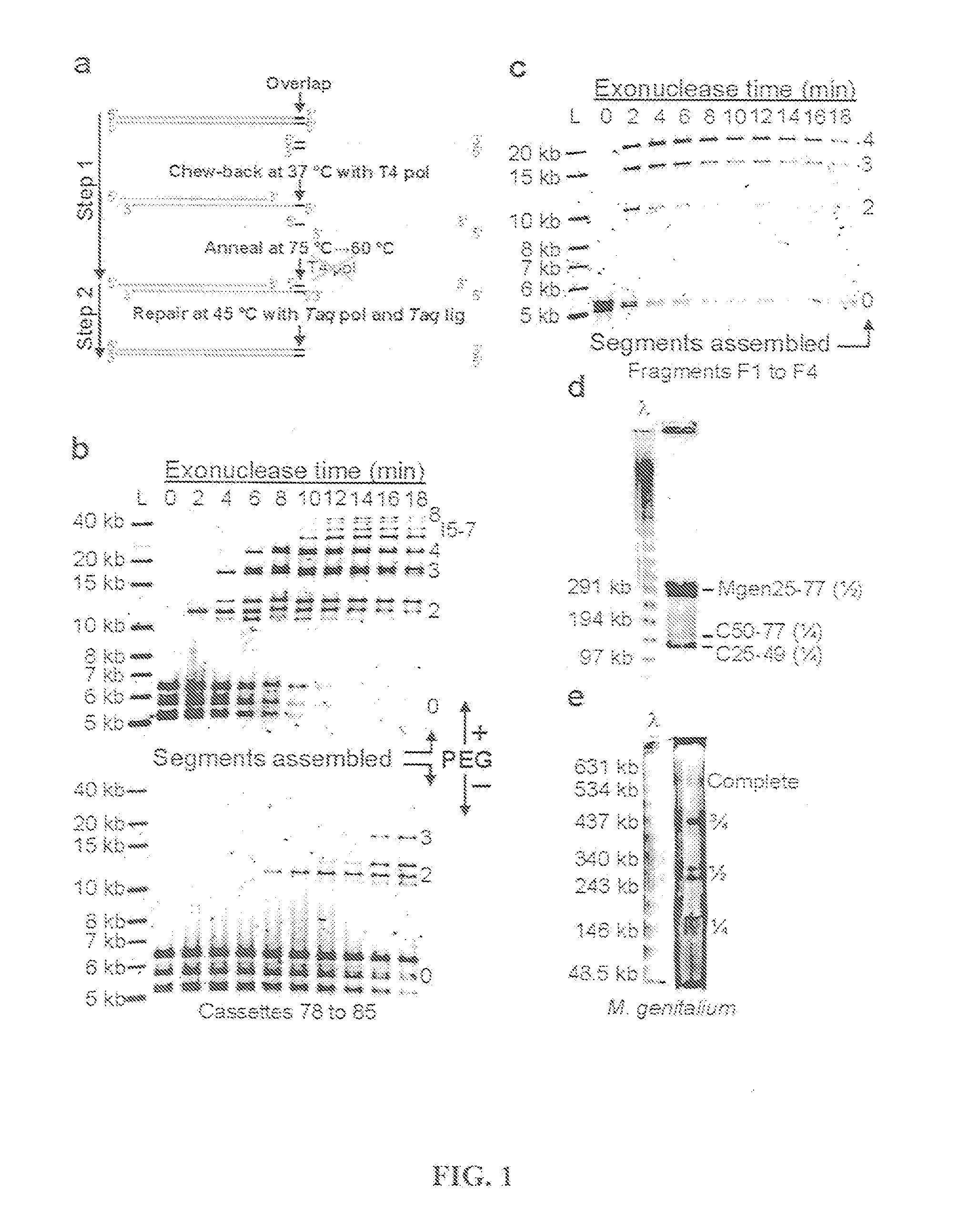Methods for in vitro joining and combinatorial assembly of nucleic acid molecules
a nucleic acid molecule and in vitro joining technology, applied in the direction of nucleotide libraries, transferases, ligases, etc., can solve the problem that no suggestion is made of using these techniques systematically to assemble the desired nucleic acid molecul
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
examples
[0208]In the following examples, all temperatures are set forth in uncorrected degrees Celsius; and, unless otherwise indicated, all parts and percentages are by weight.
example i
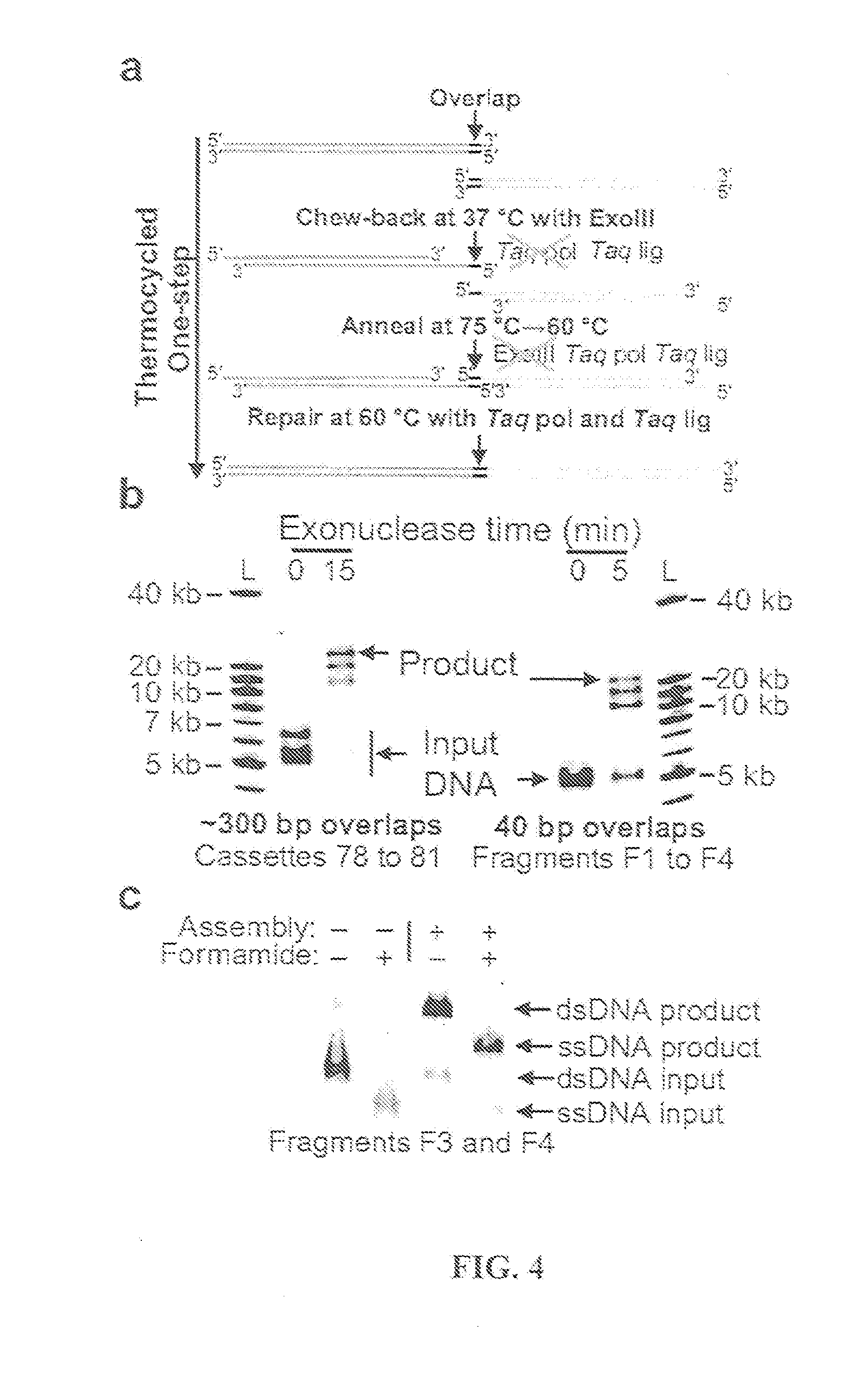
Thermocycled Exonuclease III One-Step Assembly System Protocol
[0209]A thermocycled one-step assembly was developed based on the use of an isolated non-thermostable 3′ to 5′ exonuclease and an isolated heat-activated DNA polymerase as follows. A reaction was set up on ice in a 0.2 ml PCR tube containing the following: 100 ng each substrate DNA to be assembled, 20 μl of 4×CBAR buffer, 0.7 μl of exonuclease III (4 U / μl, NEB), 8.0 μl of Taq DNA ligase (40 U / μl, NEB), 0.5 μl of AmpliTaq® Gold (5 U / μl, Applied Biosystems), and water to 80 μl. The 4×CBAR (Chew-back, Anneal, and Repair) Buffer was 20% PEG-8000, 600 mM Tris-Cl, 40 mM MgCl2, 40 mM DTT, 4 mM NAD, and 800 μM each dNTP (pH 7.5).
[0210]This gave a final concentration of 1.25 ng / μl of each DNA that was to be assembled, 5% PEG-8000, 150 mM Tris-Cl pH 7.5, 10 mM MgCl2, 10 mM DTT, 200 μM each dNTP, 1 mM NAD, 0.035 U / μl exonuclease III, 4 U / μl Taq DNA ligase, and 0.03 U / μl AmpliTaq® Gold.
[0211]100 ng substrate DNA was found to be ideal...
example ii
Isothermal T5 Exonuclease One-Step Assembly Protocol for Nucleic Acids
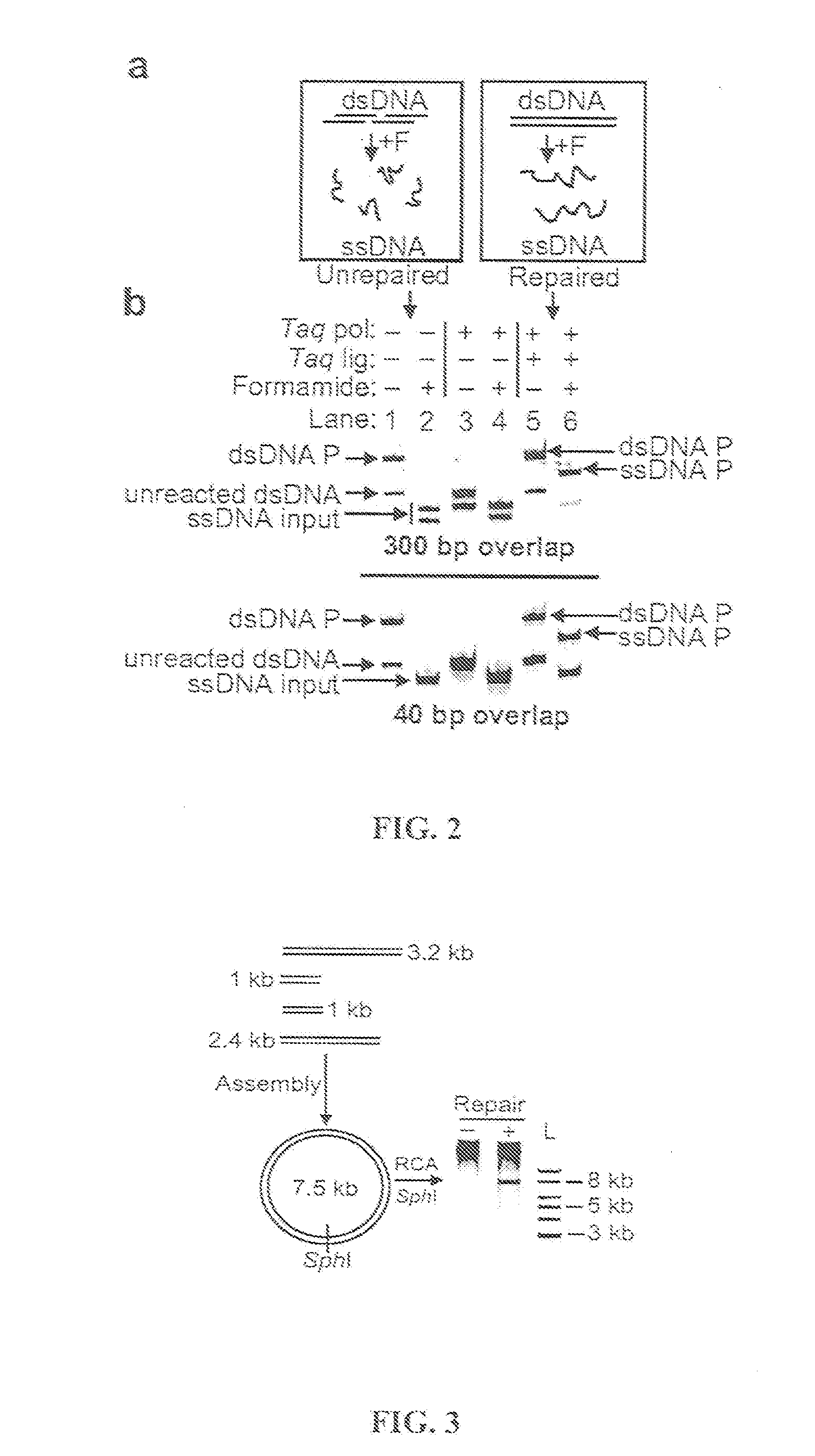
[0214]An isothermal one-step assembly was developed based on the use of an isolated non-thermostable 5′ to 3′ exonuclease that lacks 3′ exonuclease activity as follows. A reaction was set up containing the following: 100 fmol each dsDNA substrate or 3.6 μmol each ssDNA to be assembled, 16 μl 5×ISO buffer, 16 μl T5 exonuclease (0.2 U / μl, Epicentre), 8.0 μl Taq DNA ligase (40 U / μl, NEB), 1.0 μl Phusion™ DNA polymerase (2 U / μl, NEB), and water to 80 μl. The 5×ISO (ISOthermal) buffer was 25% PEG-8000, 500 mM Tris-Cl, 50 mM MgCl2, 50 mM DTT, 5 mM NAD, and 1000 μM each dNTP (pH 7.5).
[0215]This gave a final concentration of 1.25 fmol / μl each dsDNA (or 45 fmol / μl each ssDNA) that was to be assembled, 5% PEG-8000, 100 mM Tris-Cl pH 7.5, 10 mM MgCl2, 10 mM DTT, 200 μM each dNTP, 1 mM NAD, 0.02 U / μl T5 exonuclease, 4 U / μl Taq DNA ligase, and 0.03 U / μl Phusion™ DNA polymerase.
[0216]Methods used 1.64 μl 0.2 U / μl T5 exonuclease...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com