Nucleic acid construct for endogenously expressing RNA polymerase in cells
A technology of nucleic acid constructs and constructs, applied in the direction of recombinant DNA technology, enzymes, microorganisms, etc., can solve problems such as impact and bad
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0090] In the present invention, the preparation method of the yeast cell extract is not limited, and a preferred preparation method includes the following steps:
[0091] (i) providing yeast cells;
[0092] (ii) washing the yeast cells to obtain washed yeast cells;
[0093] (iii) subjecting the washed yeast cells to destructive treatment to obtain crude yeast extract;
[0094] (iv) performing solid-liquid separation on the crude yeast extract to obtain the liquid part, which is the yeast cell extract.
[0095] In the present invention, the solid-liquid separation method is not particularly limited, and a preferred method is centrifugation.
[0096] In a preferred embodiment, said centrifugation is performed in a liquid state.
[0097] In the present invention, the centrifugation conditions are not particularly limited, and a preferred centrifugation condition is 5000-100000×g, preferably 8000-30000×g.
[0098] In the present invention, the centrifugation time is not parti...
Embodiment 1
[0158] The selection of the insertion site in the cell genome of embodiment 1
[0159] In order to overcome the defect of exogenous manual addition of T7 RNA polymerase protein in the existing in vitro translation system, the present invention integrates T7 RNAP protein into the cell genome through gene editing technology, and creates a strain that can stably express T7 RNAP protein in an appropriate amount. Thus, a simple and efficient in vitro translation system without exogenous manual addition of T7 RNAP was formed.
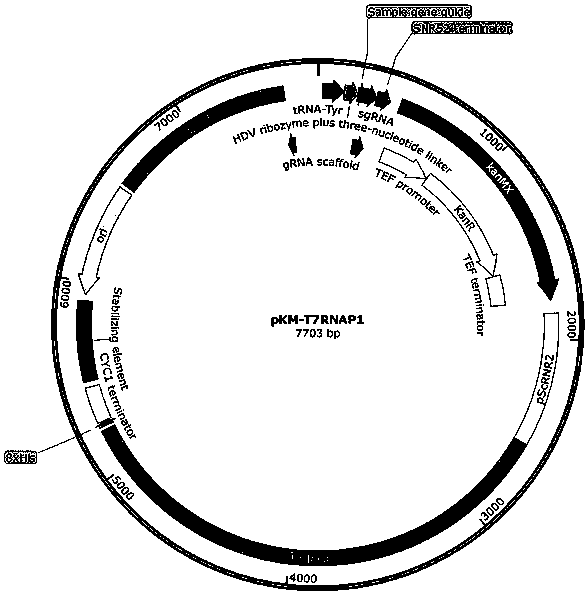
[0160] The in vitro translation system has high requirements on the content of T7 RNAP, too high or too little will affect the efficiency of the system. Therefore, the present invention inserts a weaker pScRNR2 promoter before T7RNAP, constructs the structure into an episomal plasmid, and verifies the function of the T7 RNAP expression cassette in an in vitro translation system.
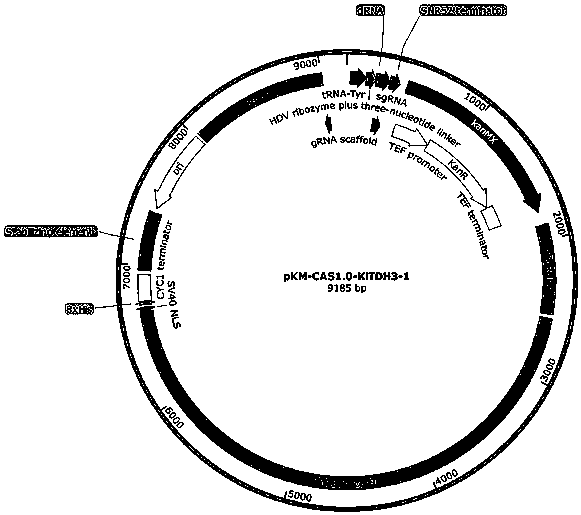
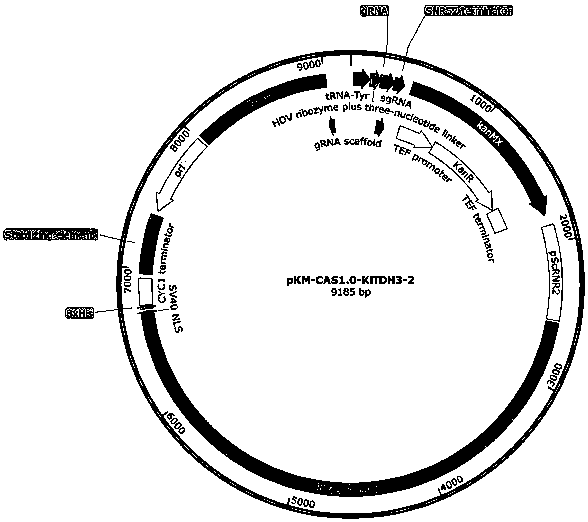
[0161] After the functional verification of the episomal plasmid, the present ...
Embodiment 2
[0162] Example 2 Construction of pKM-T7RNAP1 plasmid, yeast transformation and activity determination
[0163] 2.1 Construction of pKM-T7RNAP1 plasmid
[0164] In order to verify that T7 RNAP can be effectively expressed in cells and has transcriptional activity, the present invention first constructs T7 RNAP episomal plasmids and transforms cells. The promoter of T7 RNAP in the free plasmid is ScRNR2 promoter, the terminator is ScCYC1terminator, and the resistance gene is Kan . Plasmid construction and transformation methods are as follows:
[0165] Using the plasmid containing the T7 RNAP gene as a template, primers PF1: ATGAACACGATTAACATCGCTAAGAACG (SEQ ID NO.: 2) and PR1: TTACGCGAACGCGAAGTCCG (SEQ ID NO.: 3) were used for PCR amplification; Kluyveromyces lactis episomal plasmid was used as a template, PCR amplification was performed with primers PF2: ATCTTAGAGTCGGACTTCGCGTTCGCGTAAGAAGATGCTTCTGCTCATCATC (SEQ ID NO.: 4) and PR 2: AGTCGTTCTTAGCGATGTTAATCGTGTTCATGGTAATTGGA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com