Methods for identifying multiple DNA alteration markers in a large background of wild-type DNA
a technology of dna alteration markers and wild-type dna, which is applied in the field of methods for identifying multiple dna alteration markers in a large background of wild-type dna, can solve the problems of difficult detection of cancer, and inability to detect mutations in clinical samples, and achieves the effect of reducing the number of false positives and false positives
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
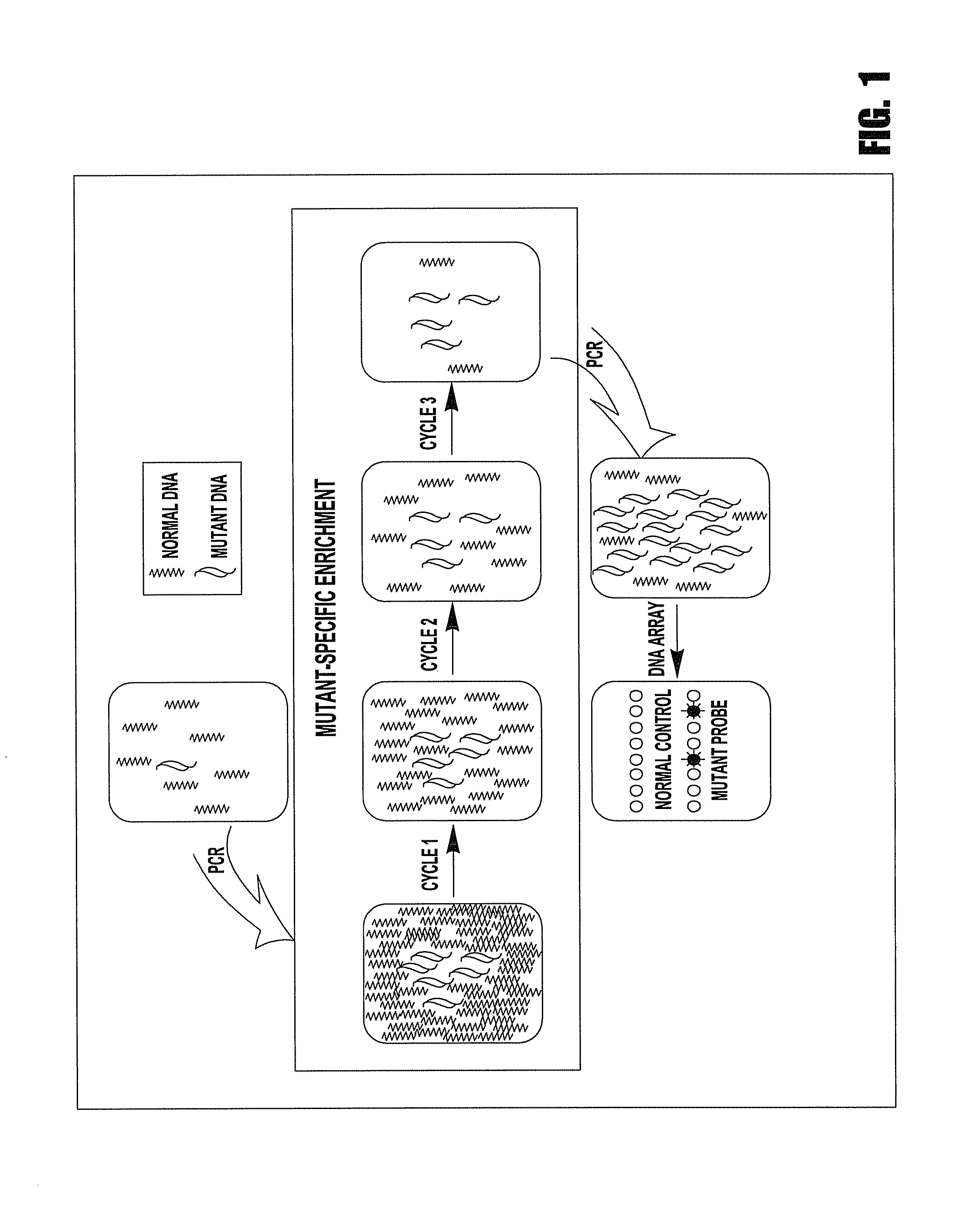
[0092]This example is to demonstrate the use of competing mutation-specific hybridization and extraction to improve the specificity of enrichment. In this method, two oligo probes were used for each mutation site, one of which (mutant probe) is complementary to mutant DNA, another of which (normal competitor probe) is complementary to wild-type DNA. However, only the mutant probes are biotinylated and thus only they and the mutant DNA carried by them can be captured, enriching mutant DNA. The use of two oligo probes is based on the consideration that the Tm change of oligos may not be sufficiently large to discriminate two alleles differing by one base and that adding a normal competitor probe can create a competition in hybridization, improving enrichment.
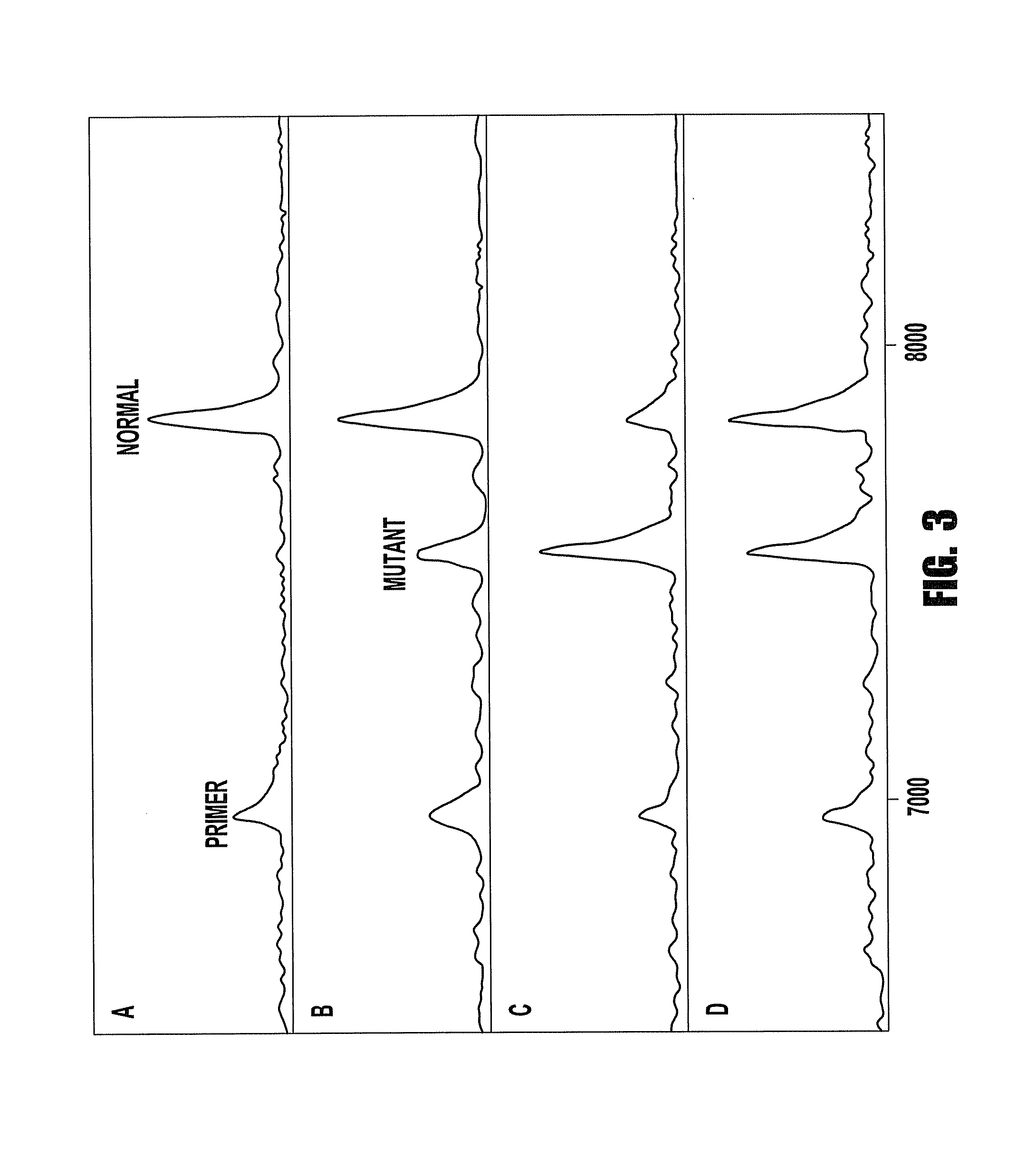
[0093]In this example, we detected a G→T mutation in the first base of codon 12 of K-ras. Wild-type DNA was obtained from Promega (Madison, Wis.). Mutant DNA was extracted from the BAL samples of the patients with lung cancer. The...
example 2
[0099]This example demonstrates the effect of the quantity of the probes used on enrichment.
[0100]In this example, we detected a G→T mutation in the first base of codon 12 of K-ras. Wild-type DNA was obtained from Promega (Madison, Wis.). Mutant DNA was extracted from the BAL samples of the patients with lung cancer. The samples containing low-abundant mutant DNA were created by diluting mutant DNA with wild-type DNA. The abundance and the quantity of mutant DNA in the created samples were estimated based on the copy number of mutant DNA in the original samples and amounts of wild-type DNA added. In this example, the sample containing 1% of mutant DNA (0.05 ng of mutant DNA in 5 ng of normal DNA) was used studied.
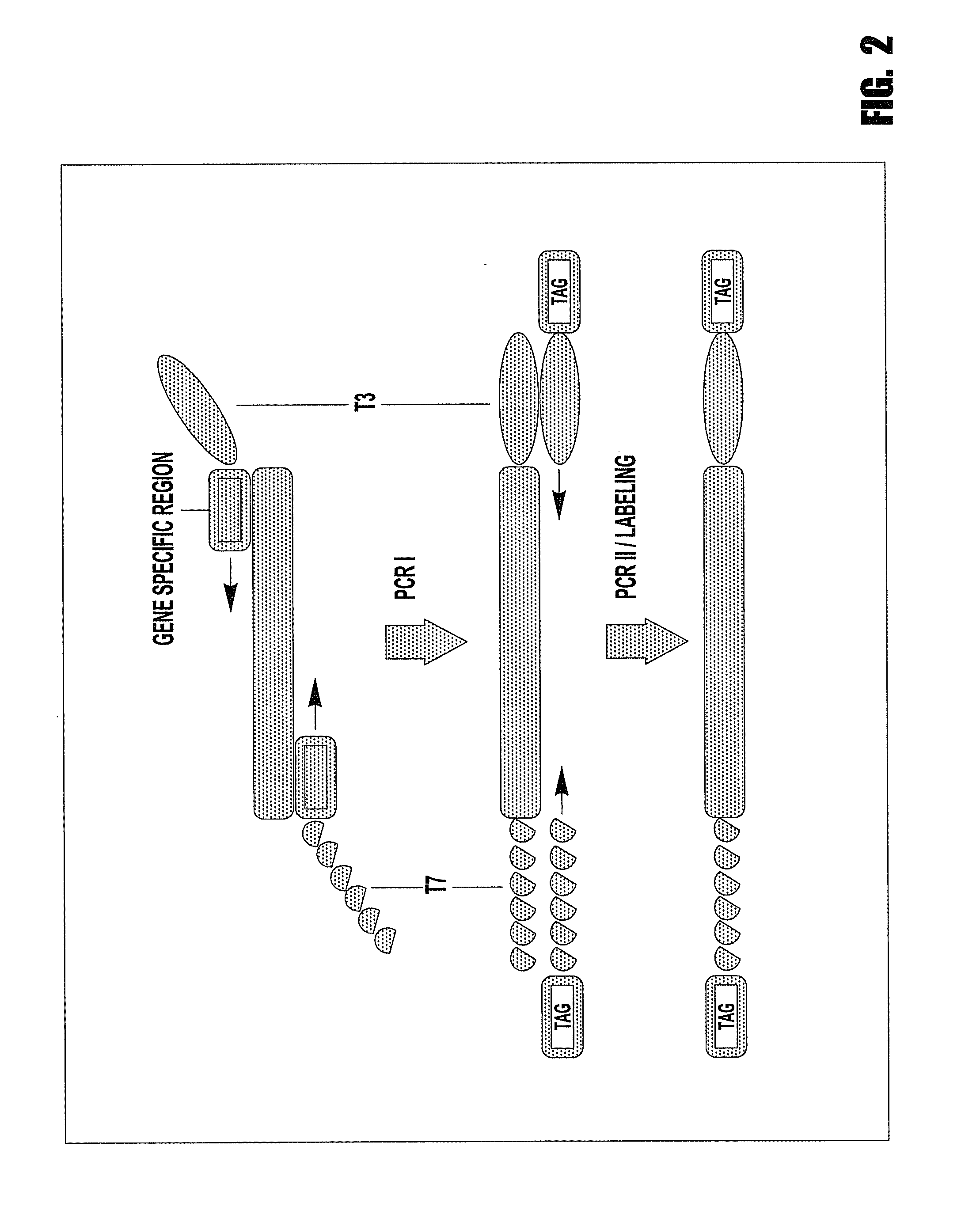
[0101]First PCR: The DNA sample was first subjected to the first PCR where forward and reverse PCR primers have a tail of either T7 or T3 and the sequences of the forward primer were: 5′-GTAATACGACTCACTATAGGAGGCCTGCTGAAAATGACTG-3′ (SEQ ID NO: 1) and the sequence of the reve...
example 3
[0106]This example is to demonstrate the improvement of enrichment using multiple cycles of hybridization and extraction. In this example, we used a sequence containing a C→T mutation on the first base of codon 1450 of APC. Wild-type DNA was obtained from Promega (Madison, Wis.). Mutant DNA was extracted from a colon cancer cell line. The samples containing low-abundant mutant DNA were created by diluting mutant DNA with wild-type DNA. The abundance and the quantity of mutant DNA in the created samples were estimated based on the copy number of mutant DNA in the original samples and amounts of wild-type DNA added. In this example, the samples containing 1%, or 0.1%, or 0.01% of mutant DNA were studied, respectively.
[0107]The forward and reverse primer of the first PCR has a tail of either T7 or T3 and their sequences were: 5′-GTAATACGACTCACTATAGGCTTCCAGATAGCCCTGGACA-3′ (SEQ ID NO: 8) and 5′-AATTAACCCTCACTAAAGGGGCAGCATTTACTGCAGCTTG-3′ (SEQ ID NO: 9), respectively. The mini-sequencing...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com