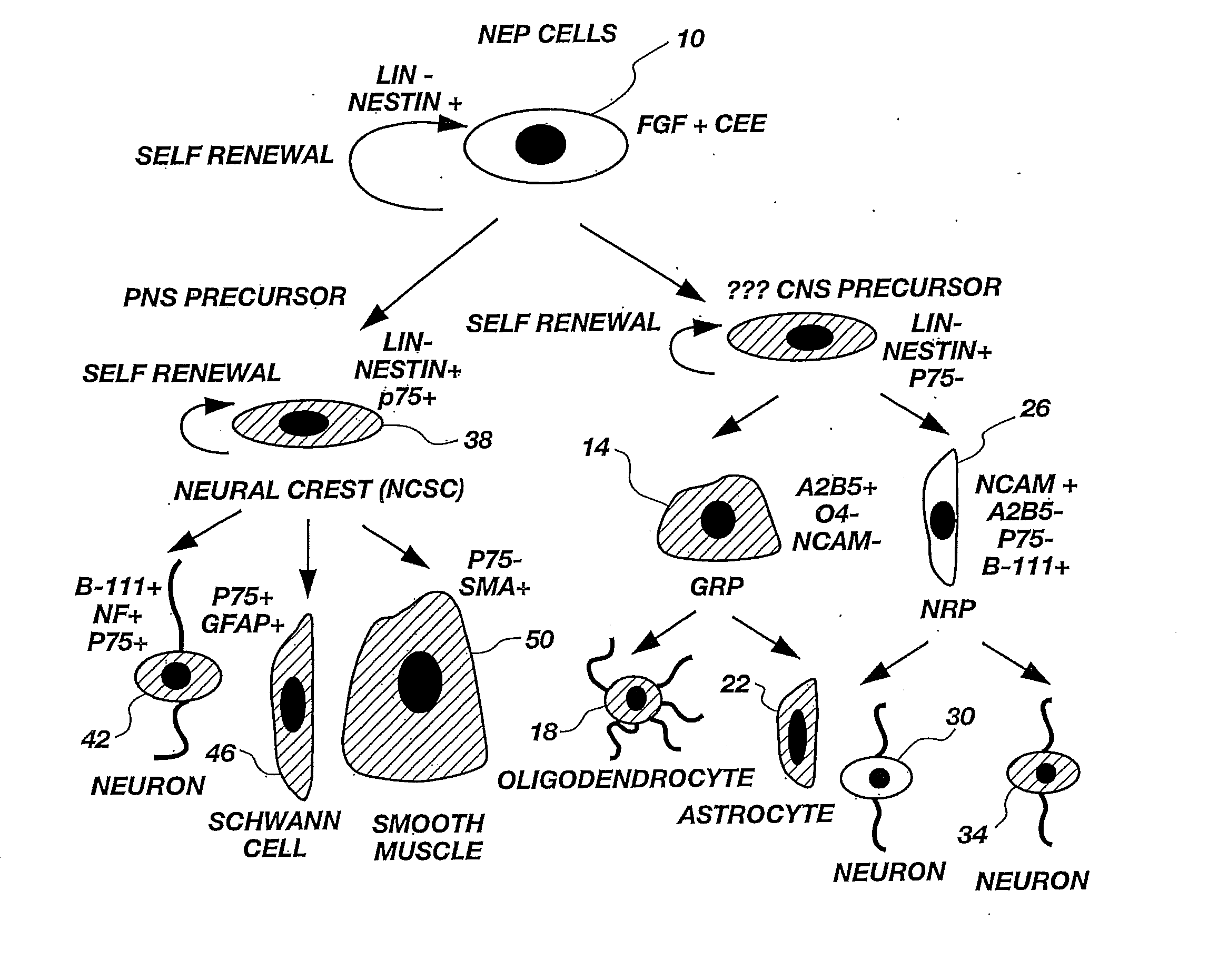
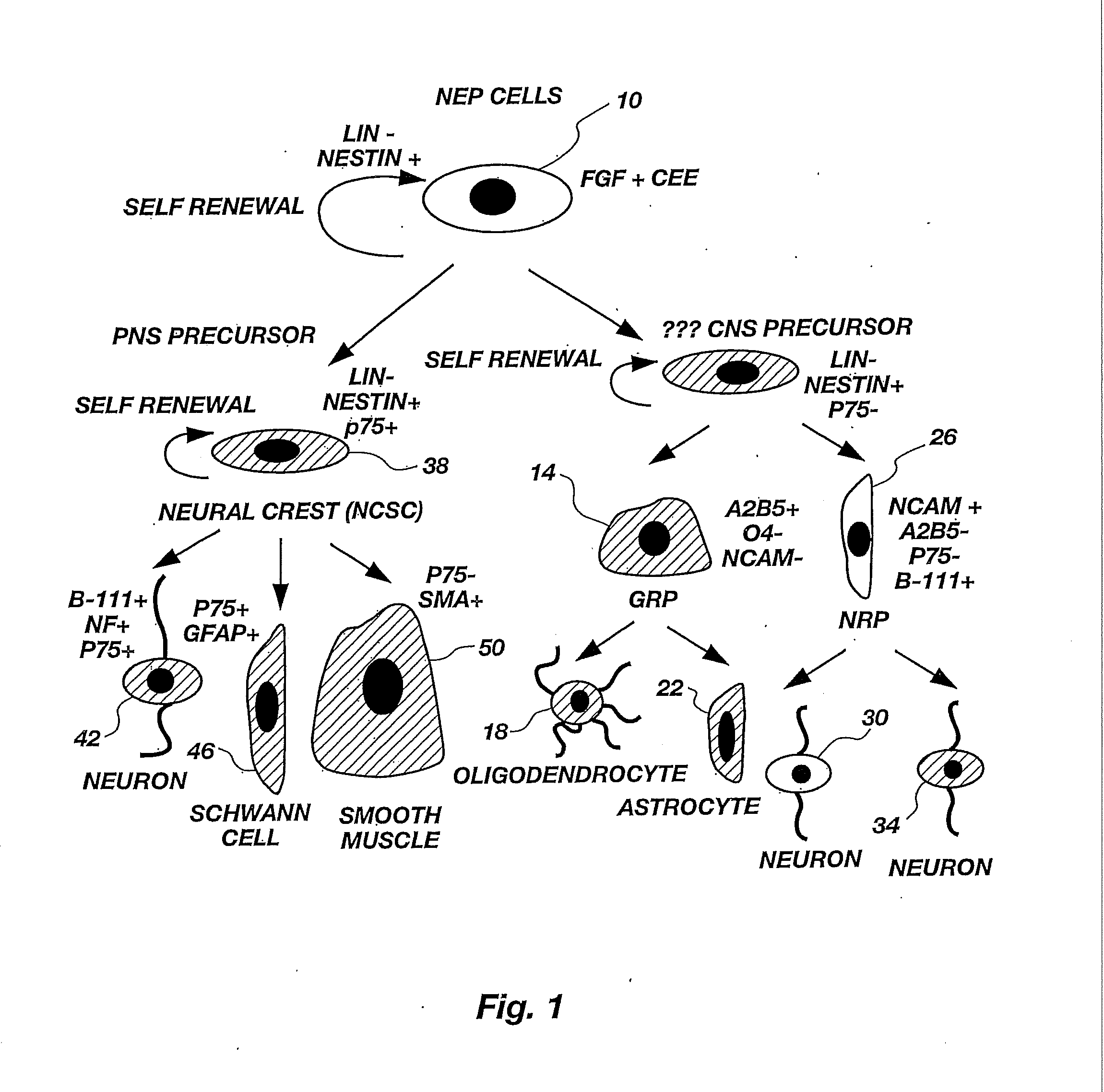
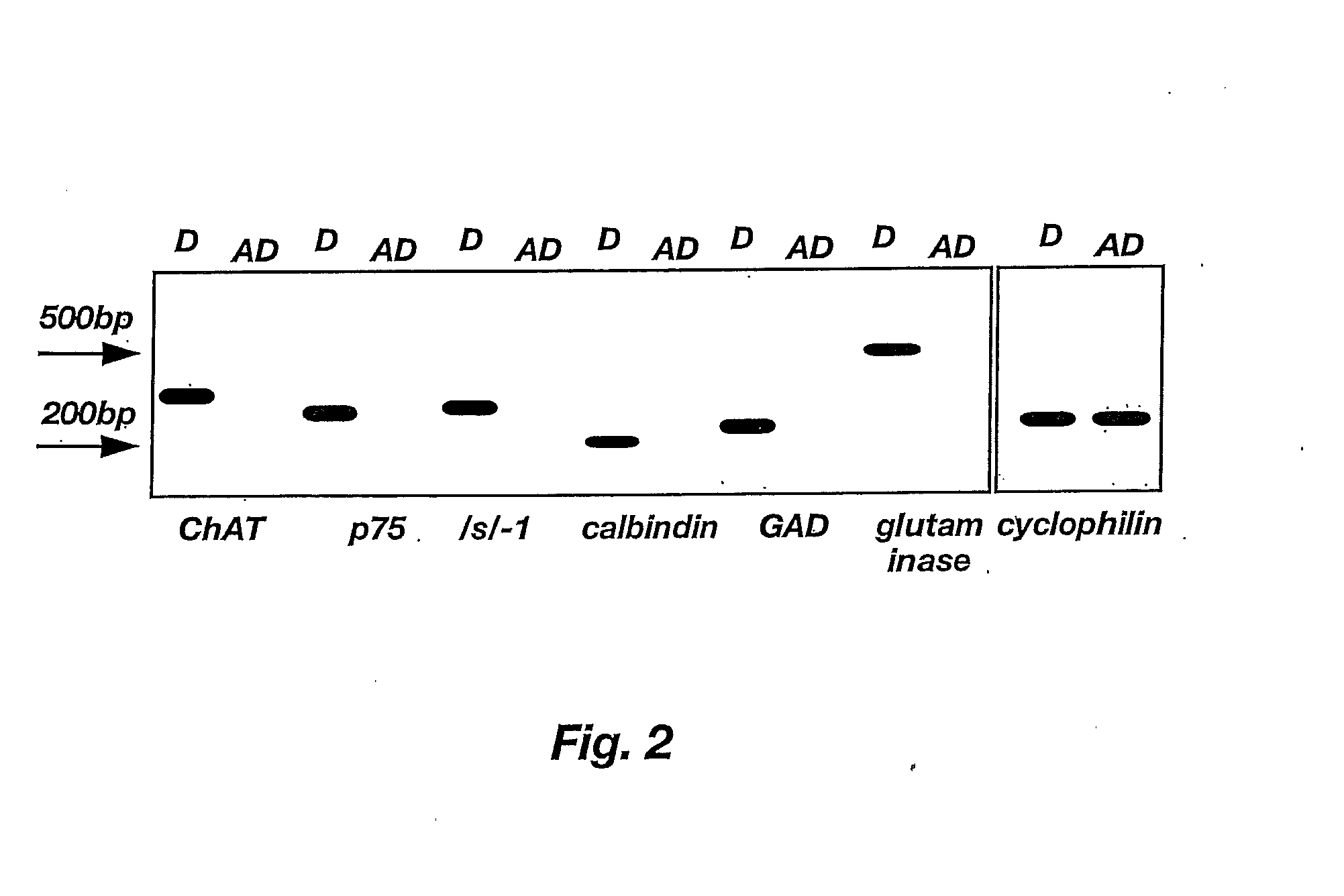
Lineage-Restricted Neuronal Precursors
a precursor cell and lineage restriction technology, applied in the field of lineage restriction intermediate precursor cells, can solve the problems of insufficient fgf failure to disclose cultured nep cells, and insufficient fgf itself for long-term growth of neurospheres,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0099]To determine if a dividing neuron-restricted precursor is normally present in vivo, sections of E13.5 rat spinal cords were analyzed with a panel of early neuronal markers. Sections were cut of embryos fresh frozen at 13.5 days gestation and then were labeled by immunocytochemistry. Staining procedures were carried out according to methods well known in the art. Cells were double-labeled with antibodies against E-NCAM (Developmental Studies Hybridoma Bank, Iowa) and β-III tubulin (Sigma Chemical Co., St. Louis, Mo.) or were stained with E-NCAM and counterstained with DAPI, a nuclear marker for identifying all cells. All secondary monoclonal antibodies were from Southern Biotechnology.
[0100]Polysialated or embryonic N-CAM (E-NCAM) appeared to be a likely marker for neuronal precursors. E-NCAM immunoreactivity was first detected at E13.5. E-NCAM immunoreactive cells could be seen in the margins of the neural tube, but not in the proliferating ventricular zone. Double-labeling wi...
example 2
[0101]To characterize E-NCAM-immunoreactive cells, E13.5 spinal cords were dissociated and E-NCAM-immunoreactive cells were stained with a panel of antibodies (Table 1). Sprague-Dawley rat embryos were removed at embryonic day 13.5 and placed in a petri dish containing Hanks balanced salt solutions (HBSS, Gibco). The trunk segments of the embryos were dissected using tungsten needles, rinsed, and then transferred to fresh HBSS. Spinal cords were mechanically dissected free from the surrounding connective tissue using sharpened No. 5 forceps. Isolated spinal cords were incubated in 0.05% trypsin / EDTA solution for 20 minutes. The trypsin solution was replaced with fresh HBSS containing 10% fetal bovine serum (FBS). The segments were gently triturated with a Pasteur pipette to dissociate cells. Cells dissociated by trituration were plated in PLL / laminin-coated 35 mm dishes (Nunc) at high density and stained after 24 hours.
TABLE 1Cell TypeAntibody / KindSourceAntigen RecognizedRecognizedA...
example 3
[0104]To determine the differentiation potential of E-NCAM-immunoreactive cells, E-NCAM+ cells were purified by immunopanning and plated at clonal density in gridded dishes. E13.5 cells were prepared according to the procedure of Example 2. An E-NCAM+ cell population was purified from these E13.5 cells using a specific antibody-capture technique according to the procedure of L. Wysocki & V. Sato, “Panning” for Lymphocytes: A Method for Cell Selection, 75 Proc. Nat'l Acad. Sci. USA 2844-48 (1978); M. Mayer et al., supra, hereby incorporated by reference. In brief, cells were trypsinized and the resulting cell suspension was plated on an A2B5-antibody-coated dish to allow binding of all A2B5+ cells to the plate. The supernate was removed, and the plate was washed with DMEM supplemented with additives described by J. Bottenstein and G. Sato, Growth of a Rat Neuroblastoma Cell Line in Serum-free Supplemented Medium, 76 Proc. Nat'l Acad. Sci. USA 514-17 (1979), hereby incorporated by ref...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com