Identification of Tumor-Associated Markers for Diagnosis and Therapy
a tumor-associated marker and tumor technology, applied in the field of nucleic acids and encoded polypeptides, can solve the problem that cancer remains among the leading causes of death
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Screening for Placenta-Specific Genes Aberrantly Activated in Tumors
[0222]Tissues and Cell Lines
[0223]Tissues were obtained as human surplus materials during routine diagnostic or therapeutic procedures and were stored at −80° C. until use. Cell lines were purchased from the American Type Culture Collection (ATCC) and the German Resource Collection of Microorganisms and Cell Culture (DSMZ).
[0224]RNA Isolation and Microarray Hybridization
[0225]Total RNA was isolated using the RNeasy Mini Kit protocol (Qiagen). Quantification of isolated RNA was performed using UV-spectroscopy and the quality was determined both by A260 / A280 ratio and Agilent bioanalyzer (Agilent Technologies). Five micrograms total RNA were used for cDNA synthesis with 5 pmol μl−1 T7-oligo(dT)24 primer and was performed at 43° C. for 90 minutes with the “Superscript First-Strand Synthesis-System” for RT-PCR (Invitrogen). Second-strand synthesis was performed with complete cDNA. The cDNA solution was incubated at 160°...
example 2
Validation of the Identified Tumor-Associated Markers
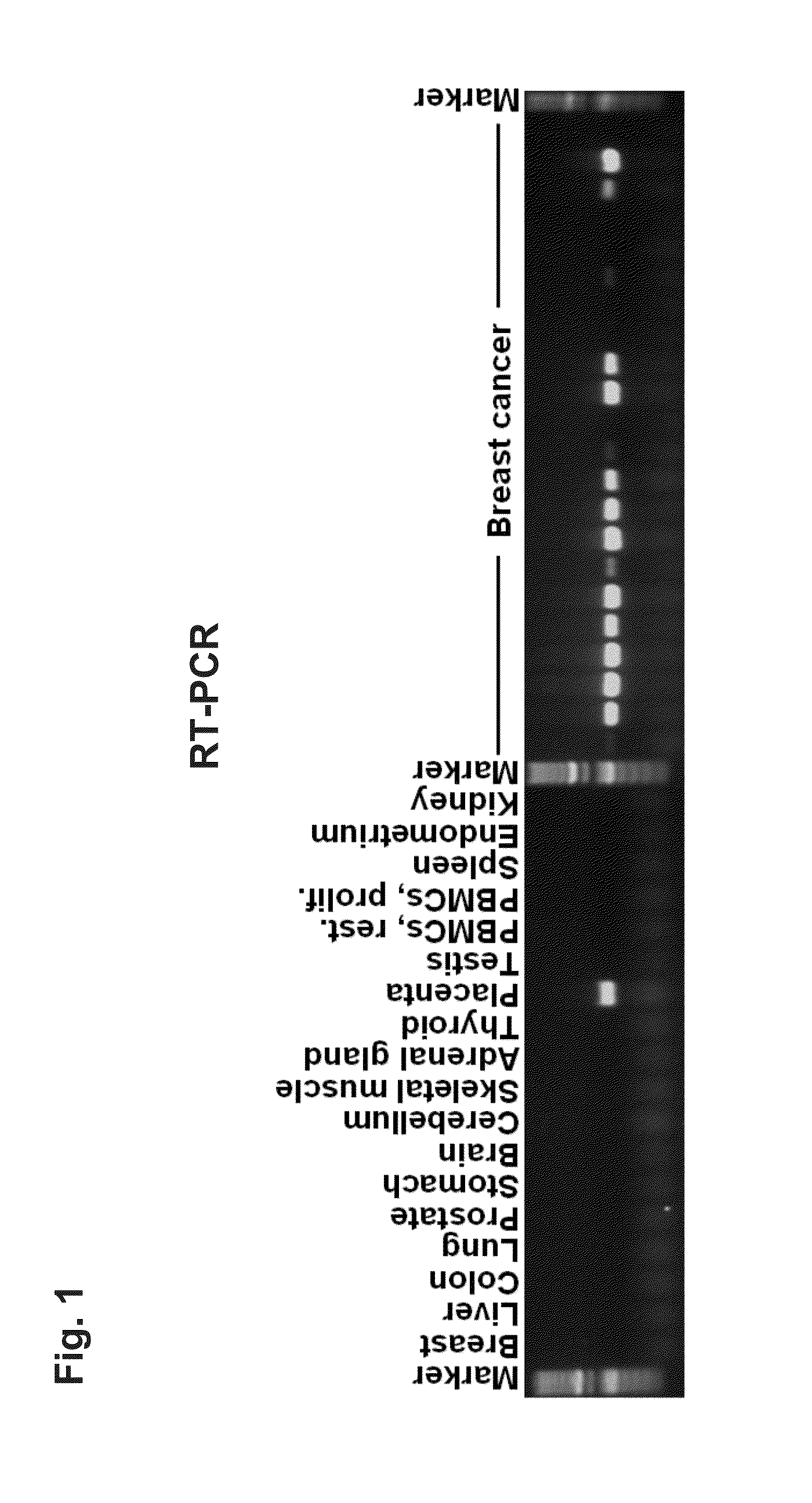
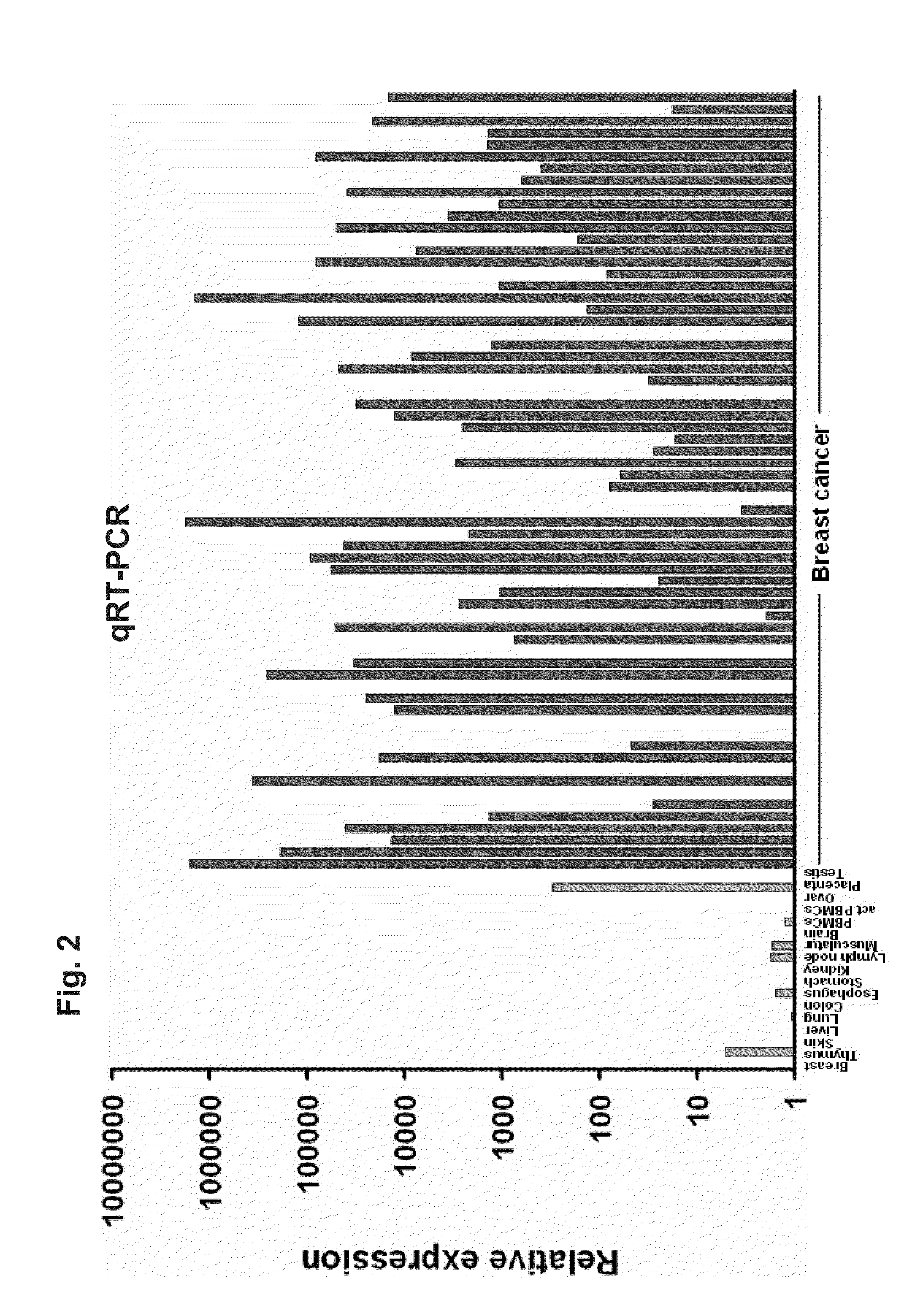
[0230]1. Examination of RNA Expression
[0231]The identified tumor-associated markers are first validated with the aid of RNA which is obtained from various tissues or from tissue-specific cell lines. Since the differential expression pattern of healthy tissue in comparison with tumor tissue is of decisive importance for the subsequent therapeutic application, the target genes are preferably characterized with the aid of these tissue samples.
[0232]Total RNA is isolated from native tissue samples or from tumor cell lines by standard methods of molecular biology. Said isolation may be carried out, for example, with the aid of the RNeasy Maxi kit (Qiagen, Cat. No. 75162) according to the manufacturer's instructions. This isolation method is based on the use of chaotropic reagent guanidinium isothiocyanate. Alternatively, acidic phenol can be used for isolation (Chomczynski & Sacchi, Anal. Biochem. 162: 156-159, 1987). After the tissue ...
example 3
Detailed Analysis of the Identified Tumor-Associated Markers
[0285]RNA-Isolation, RT-PCR and Real-Time RT-PCR
[0286]RNA extraction, first-strand cDNA synthesis, RT-PCR and real-time RT-PCR was performed as previously described (Koslowski, M. et al., Cancer Res. 62, 6750-6755 (2002), Koslowski, M. et al., Cancer Res. 64, 5988-5993 (2004)). Real-time quantitative expression analysis was performed in a 40 cycle RT-PCR. After normalization to HPRT (sense 5′-TGA CAC TGG CAA AAC AAT GCA-3′; antisense 5′-GGT CCT TTT CAC CAG CAA GCT-3′, 62° C. annealing) gene-specific transcripts in tumor samples were quantified relative to normal tissues using ΔΔCT calculation.
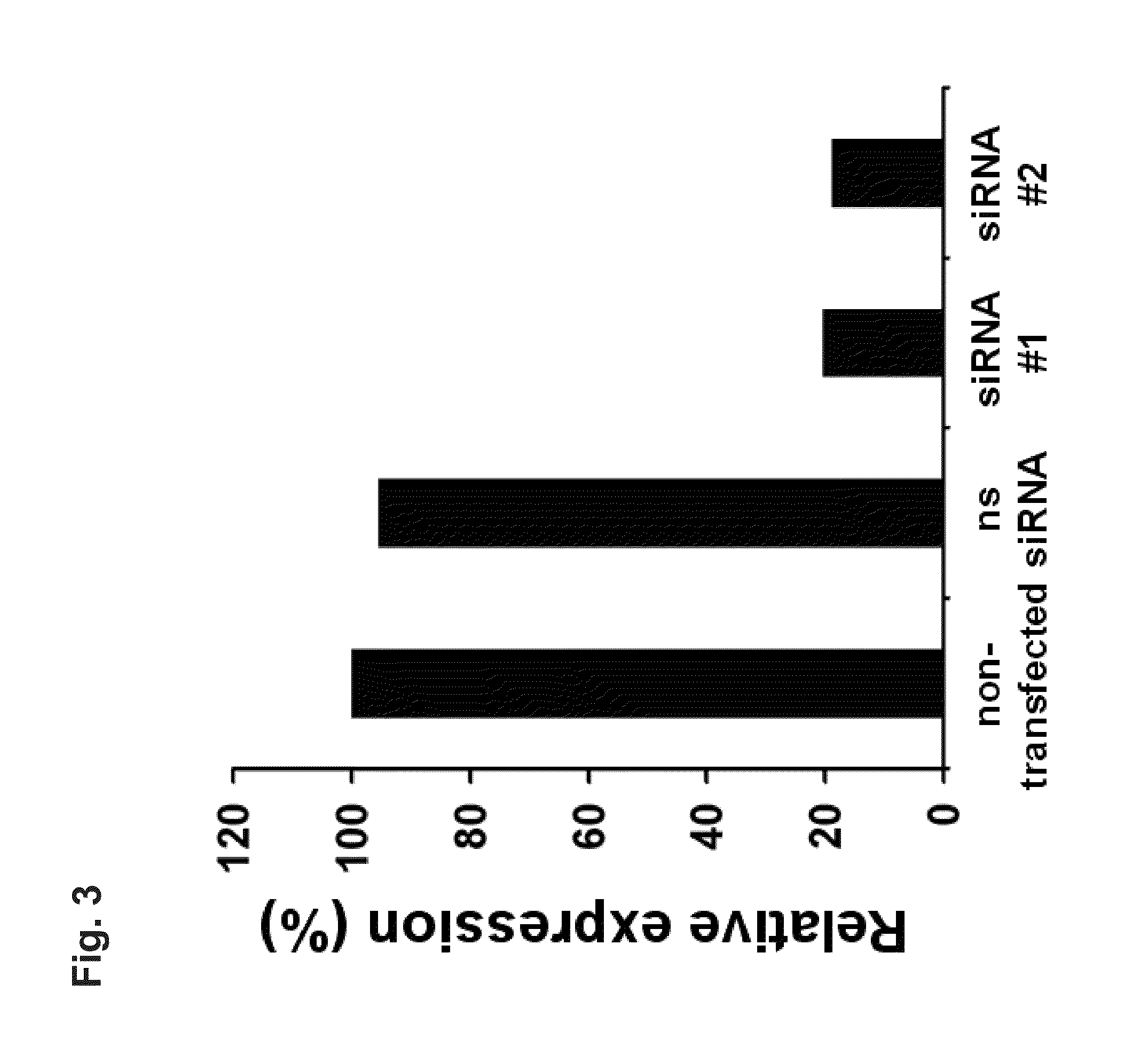
[0287]siRNA Duplexes
[0288]The SEQ ID NO:540 siRNA duplexes (Qiagen, Hilden, Germany) were directed against target sequences 5′-NNC CAC AGA AGG UAC CAG UUA-3′ (siRNA #1; sense (5′-CCA CAG AAG GUA CCA GUU AUU-3′), antisense (5′-UAA CUG GUA CCU UCU GUG GUU-3′) and 5′-NNC AGC AAG ACU CCC UCU AAA-3′ (siRNA #2; sense (5′-CAG CAA GAC UCC CUC ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pharmaceutical composition | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com