Binding proteins for recognition of DNA
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 2
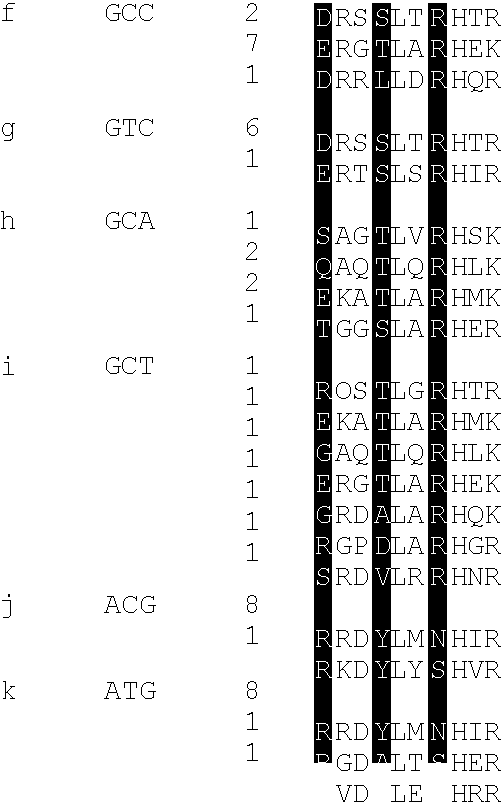
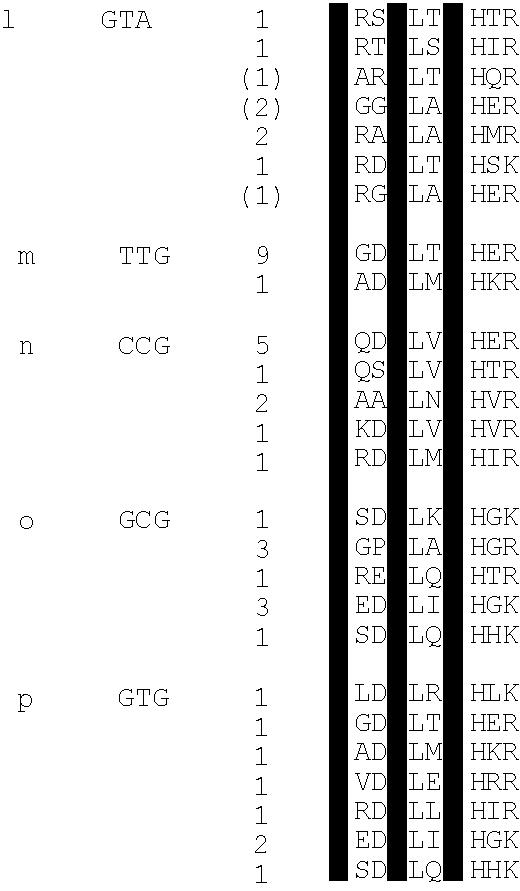
[0105]This example describes a new technique to deal efficiently with the selection of a DNA binding site for a given zinc finger (essentially the converse of example 1). This is desirable as a safeguard against spurious selections based on the screening of display libraries. This may be done by screening against libraries of DNA triplet binding sites randomised in two positions but having one base fixed in the third position. The technique is applied here to determine the specificity of fingers previously selected by phage display. The inventors found that some of these fingers are able to specify a unique base in each position of the cognate triplet. This is further illustrated by examples of fingers which can discriminate between closely related triplets as measured by their respective equilibrium dissociation constants. Comparing the amino acid sequences of fingers which specify a particular base in a triplet, we infer that in most instances, sequence specific binding of zinc fi...
example 3
[0133]From the evidence presented in the preceding examples, the inventors propose that specific DNA-binding proteins comprising zinc fingers can be “made to measure”. To demonstrate their potential the inventors have created a three finger polypeptide able to bind site-specifically to a unique 9 bp region of a BCR-ABL fusion oncogene and to discriminate it from the parent genomic sequences (Kurzrock et al., 1988 N. Engl. J. Med. 319, 990 -988). Using transformed cells in culture as a model, it is shown that binding to the target oncogene in chromosomal DNA is possible, resulting in blockage of transcription. Consequently, murine cells made growth factor-independent by the action of the oncogene (Daley et al., 1988 Proc. Natl. Acad. Sci. U.S.A. 85, 9312-9316) are found to revert to factor dependence on transient transfection with a vector expressing the designed zinc finger polypeptide.
[0134]DNA-binding proteins designed to recognise specific DNA sequences could be incorporated in c...
example 4
[0152]The phage display zinc finger library described in the preceding examples could be considered sub-optimal in a number of ways:
[0153]i) the library was much smaller than the theoretical maximum size;
[0154]ii) the flanking fingers both recognised GCG triplets (in certain cases creating nearly symmetrical binding sites for the three zinc fingers, which enables the peptide to bind to the ‘bottom’ strand of DNA, thus evading the register of interactions we wished to set);
[0155]iii) Asp+2 finger three (“Asp++2”) was dominant over the interactions of finger two (position+6) with the 5′ base of the middle triplet;
[0156]iv) not all amino acids were represented in the randomised positions.
[0157]In order to overcome these problems a new three-finger library was created in which:
[0158]a) the middle finger is fully randomised in only four positions (−1, +2, +3 and +6) so that the library size is smaller and all codons are represented. The library was cloned in the pCANTAB5E phagemid vector...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Affinity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com