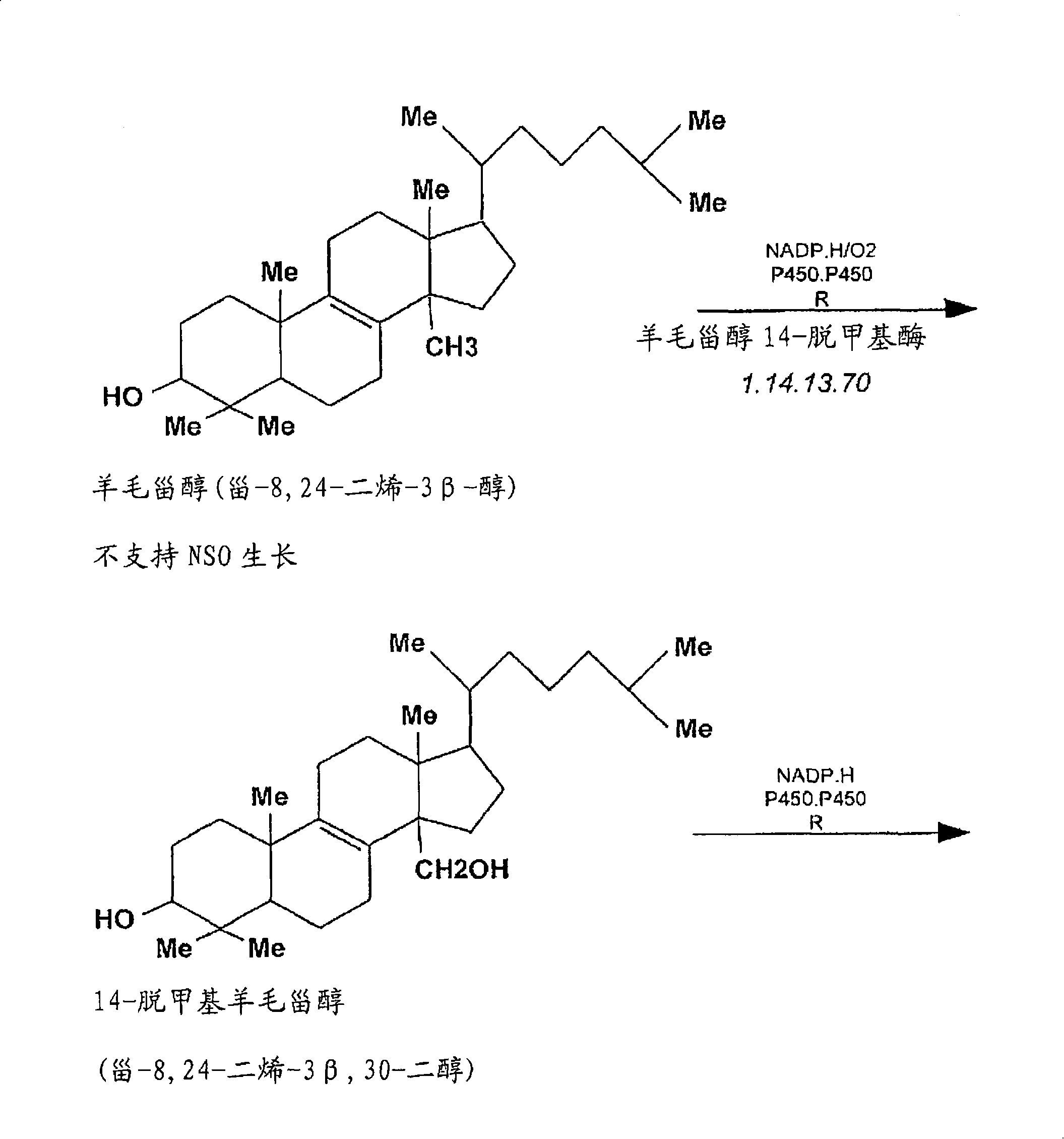
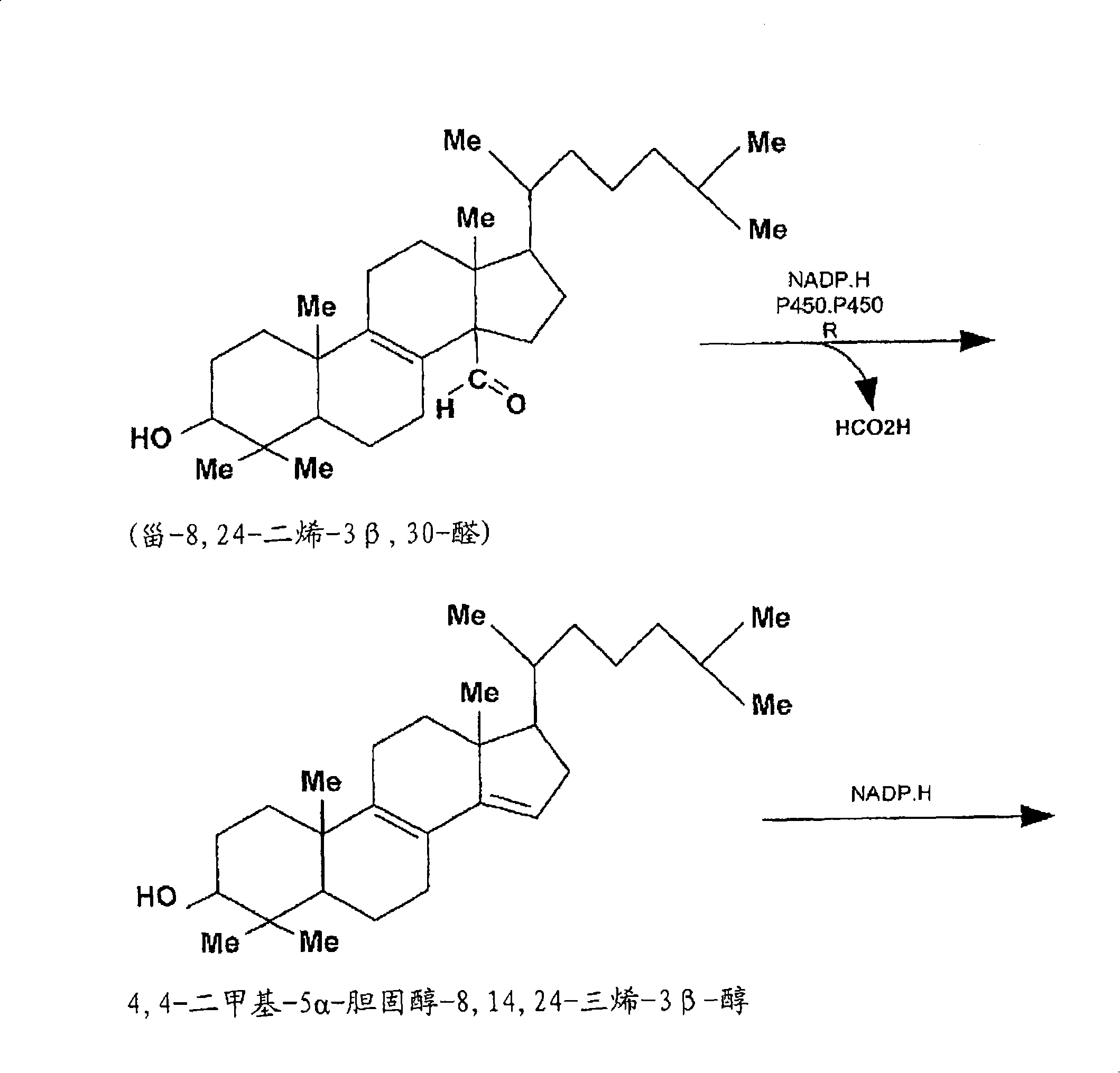
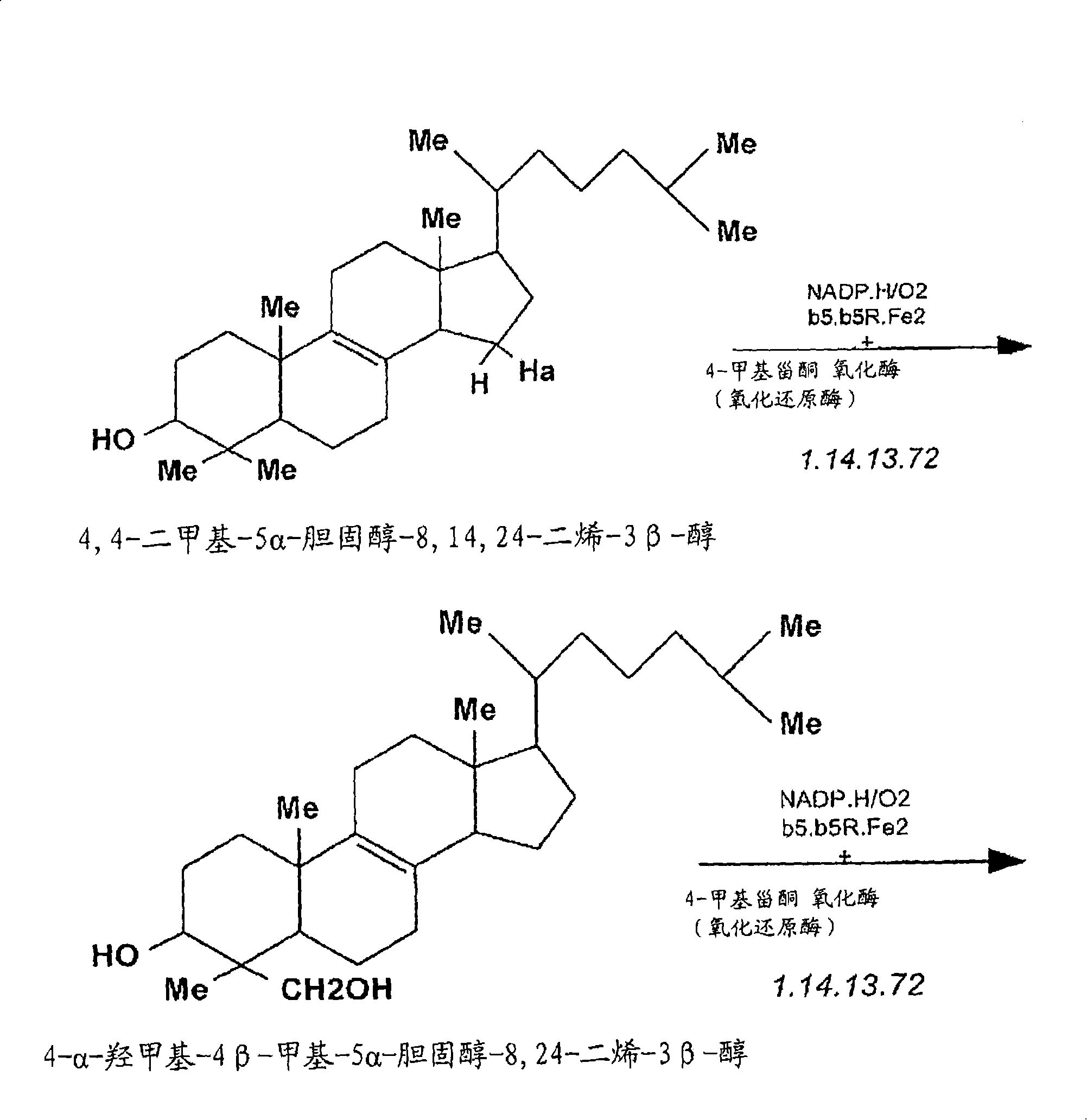
Compositions and methods for metabolic selection of transfected cells
A cell and cell sterol technology, which is applied in the field of compositions and methods for metabolic selection of transfected cells, can solve the problems of unstable coupling process, easy occurrence, and difficulty in filtering through small-pore sterile membranes.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0160] Example 1: Elaboration of Cholesterol Screening of Mouse Myeloma Cell Line NS-0 in Chemically Defined or / and Serum-Free Cell Culture Medium
[0161] The mouse myeloma cell line NS-0 was adapted in two chemically defined serum-free cell culture media to test the suitability of 3-KSR as a selectable marker in this cellular background or under SFM conditions. Using CD-hybridoma ( ) and CDM-4-NS-0 ( ) for adaptive culture of NS-0 cells. NS-0 cell lines from each adapted culture in CD-SFM were used to establish the Accession cell bank. Establish a method that can be used to determine the dependence of CD-SFM-NS-0 cell line on cholesterol in cell culture medium. The results of these experiments indicated that less than 10% of CD-SFM-NS-0 cells could survive after 72 hours of culture in cholesterol-free medium. After an additional 5 days of culture in CD-SFM, no viable cells were detected using trypan blue staining. In addition, in the absence of cholesterol, no intermi...
Embodiment 2
[0162] Embodiment 2: the PCR reaction of mouse 3-ketosterol reductase (3-KSR) and construction 3-KSR expression vector
[0163] PCR amplification of the 3-ketosterol reductase (3-KSR) gene encoding the mouse muscuhis 3B-hydroxy-delta(5)-steroid deseroid was performed using cDNA isolated from adult male BALB / c mouse kidney Hydrogenase (Hsd3b5). Using the mouse 3-KSRHsd3b5 sequence reported in the references, using its specific oligonucleotide sequence for PCR amplification reaction, a 1.1 kb band can be detected in agarose gel electrophoresis. This band was isolated and cloned into the pCR-Blunt II-TOPO vector ( , and then re-cloned into the pBV dhfr.1 plasmid background, the new building block vector is called pBFksr.1.
[0164] The 1.1kb region encoding 3-ksr in pBFksr.1 was confirmed by DNA sequencing, and its open reading frame (orf) was compared with the published sequence, and the sequencing results were consistent with the reported mouse 3-ketosterol reductase ( NCBI...
Embodiment 3
[0165] Example 3: Transfection and selection of NS-0 cells in chemically defined fatty acid-free selection medium
[0166] A pBF / csr.1 containing the correct 3-KSR ORF was transfected into NS-0 cells and selected in chemically defined fatty acid-free selection medium. The cell selection medium includes the following components: CD-hybridoma ( ), Glutamax (2mM, ), NEAA (non-essential amino acids) (Ix, ), fatty acid-free BSA (1%, Calbiochem), recombinant human IL-6 (5ng / ml, Promega), ITS broth supplement (Ix, Sigma-Aldrich). Initial transfection and selection can be performed in T-75 cell culture flasks. On the day of transfection, NS-0 cells were counted using trypan blue staining to differentiate live or dead cells. At least 90% of the cells survived. Approximately 1 x 10 7 . In addition to plasmid transfection, a negative control transfection (containing no DNA) needs to be performed. The cells were centrifuged, resuspended in 20 ml of serum-free transfection mediu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com